Metatranscriptomic Analysis of Argentinian Kefirs Varying in Apparent Viscosity
Abstract
1. Introduction
2. Materials and Methods
2.1. Kefir Grains and Fermented Milk (kefir) Preparation
2.2. Physicochemical and Microbiological Characterisation of Fermented Milk
2.3. Identification of the Transcriptionally Active Microorganisms in Kefir by RNA-seq Analysis
2.4. Bioinformatic Analysis
3. Results and Discussion
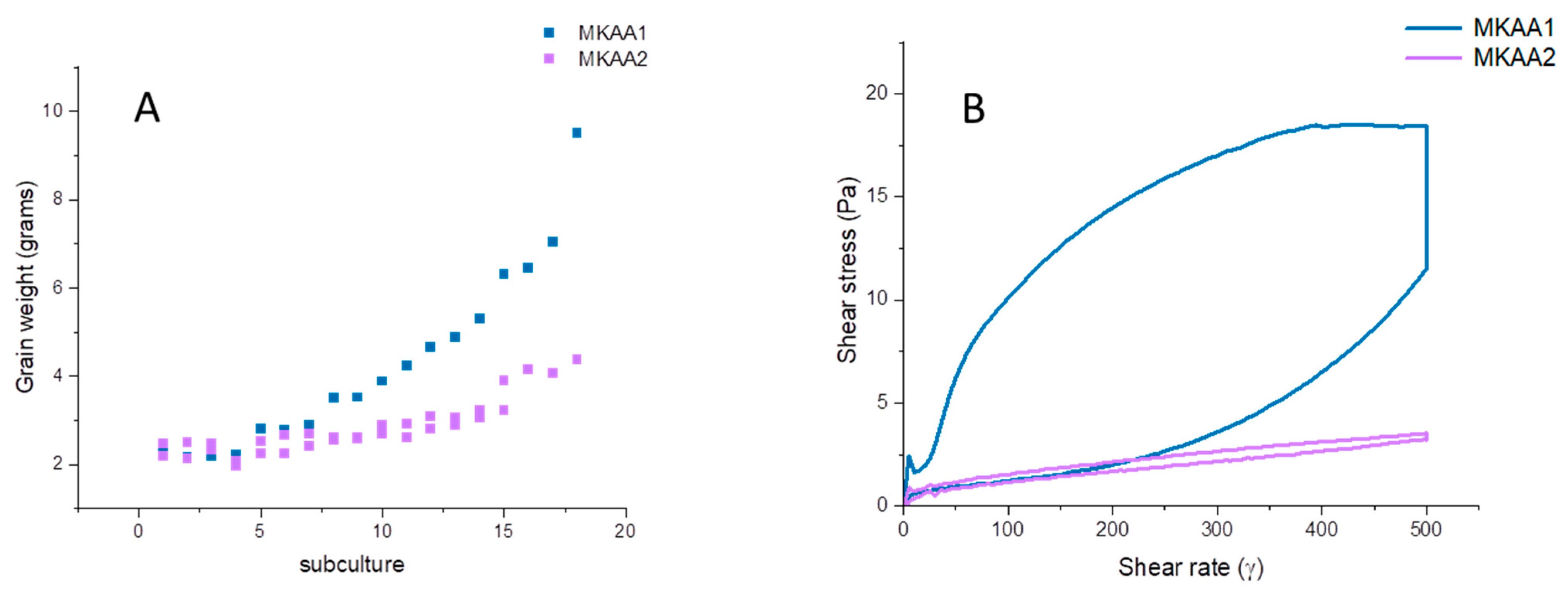
3.1. Kefir Biomass Growth during Successive Subcultures and Physicochemical and Microbiological Characterisation of Kefir
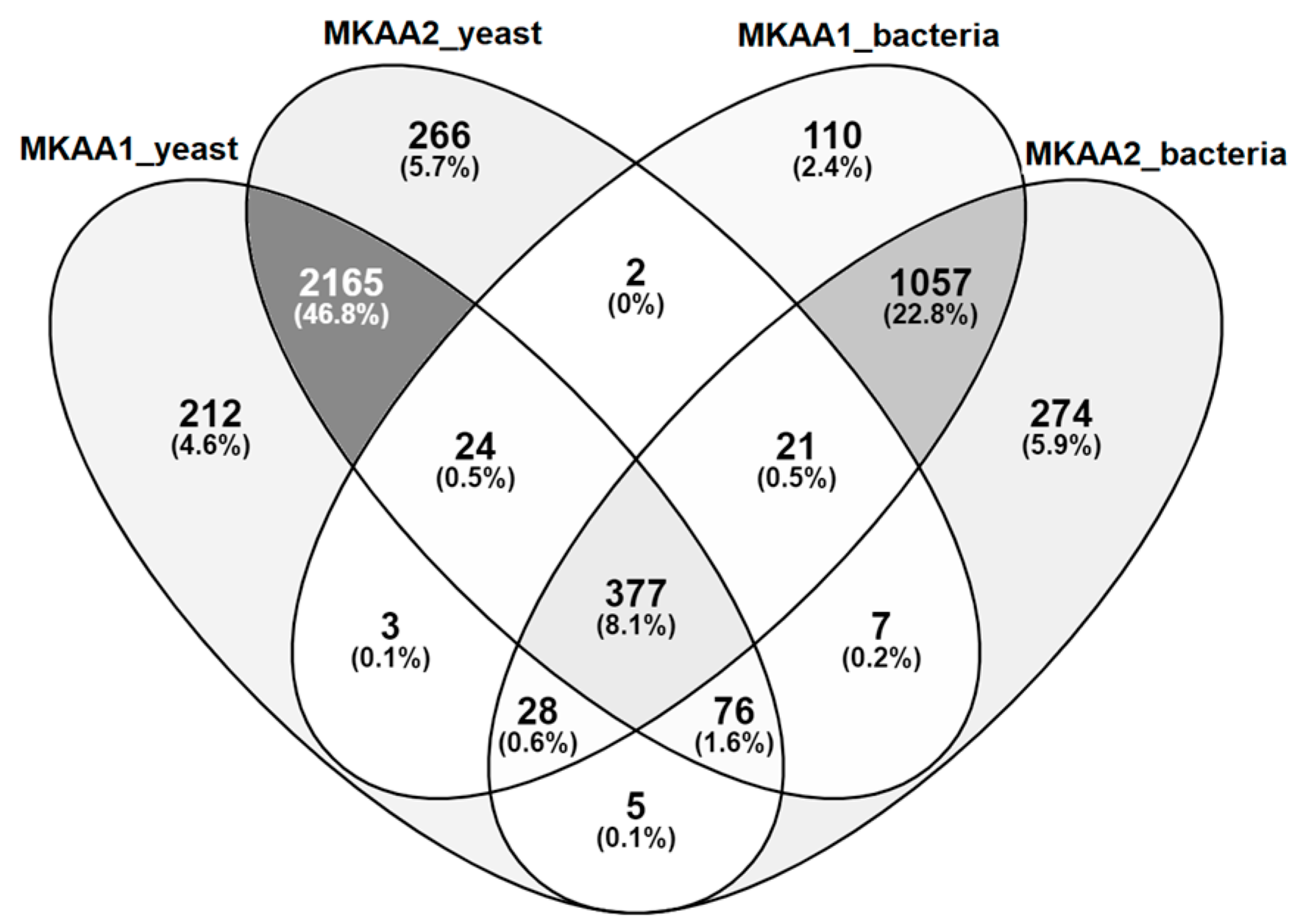
3.2. Sequencing Overview
3.3. The Transcriptionally Active Microbiome (TAM) of MKAA1 and MKAA2 Kefir
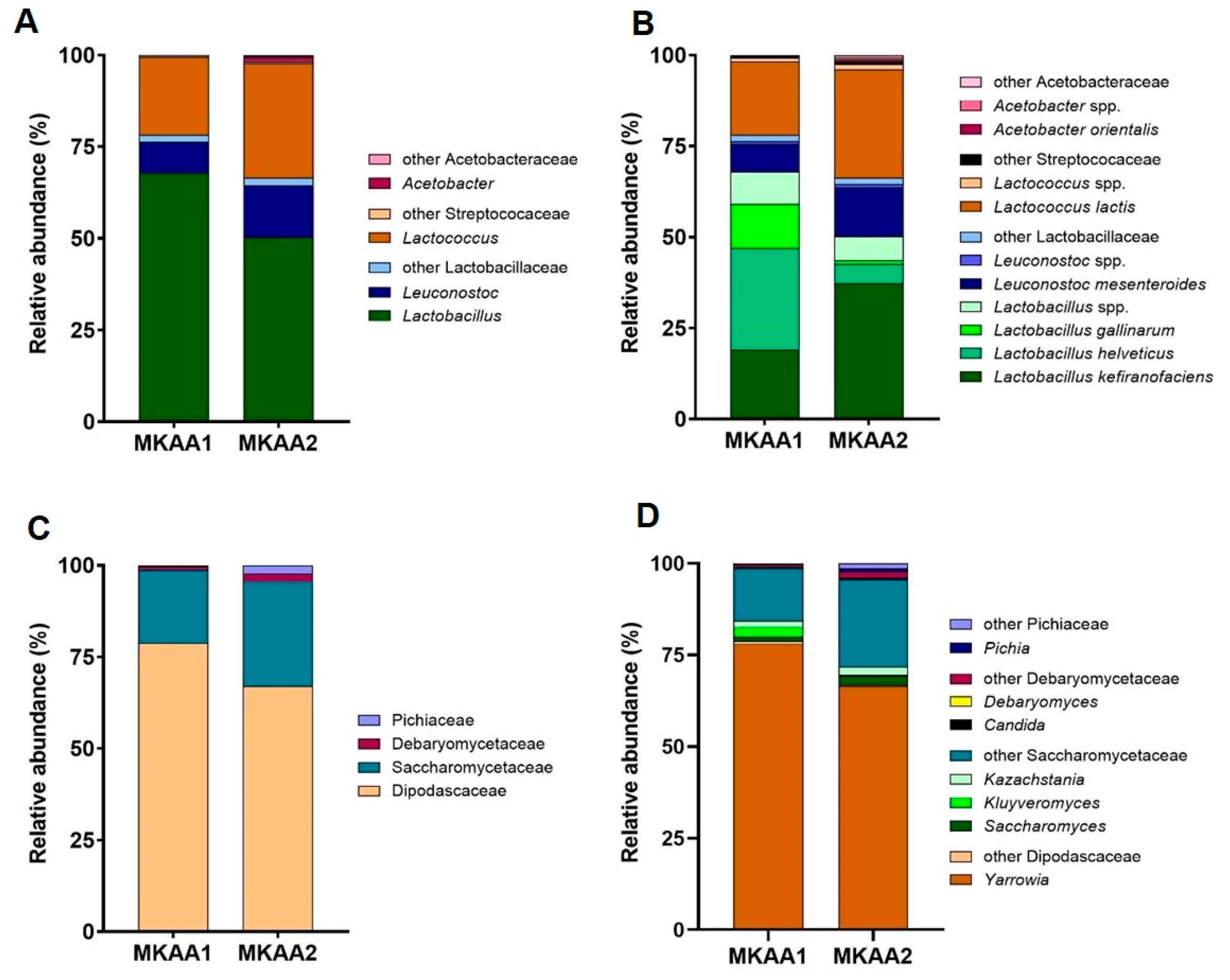
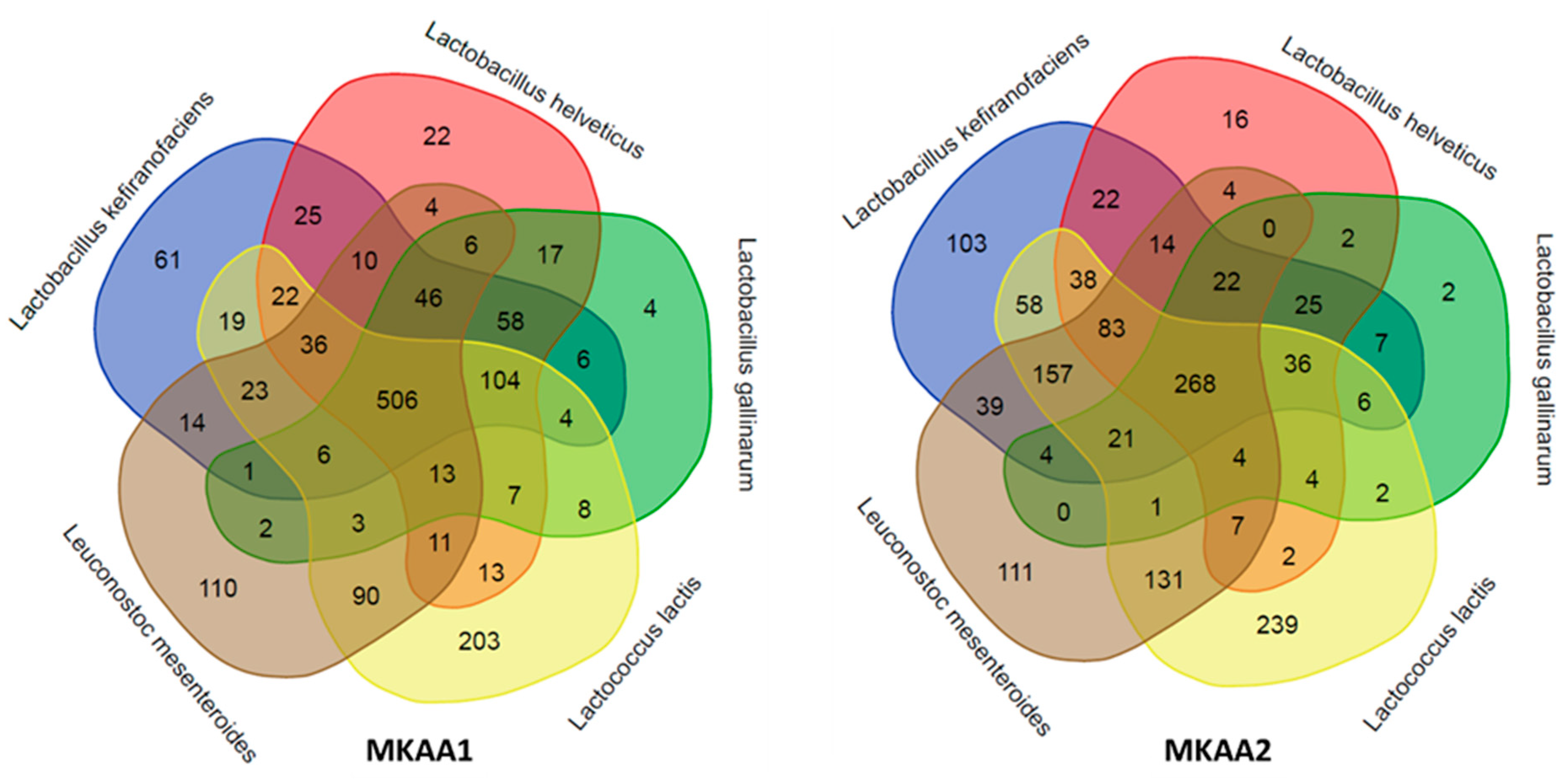
3.3.1. Bacteria Taxonomy in the Metatranscriptome of Kefir Beverages
3.3.2. Yeast Taxonomy in the Metatranscriptome of Kefir Beverages
3.4. The Functional Profile of the Kefir Microbial Community of MKAA1 and MKAA2 Beverages
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leite, A.M.O.; Miguel, M.A.L.; Peixoto, R.S.; Rosado, A.S.; Silva, J.T.; Paschoalin, V.M.F. Microbiological, Technological and Therapeutic Properties of Kefir: A Natural Probiotic Beverage. Braz. J. Microbiol. 2013, 44, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Osimani, A.; Milanović, V.; Aquilanti, L.; De Filippis, F.; Stellato, G.; Di Mauro, S.; Turchetti, B.; Buzzini, P.; Ercolini, D.; et al. Bacteria and Yeast Microbiota in Milk Kefir Grains from Different Italian Regions. Food Microbiol. 2015, 49, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Fiorda, F.A.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; Pagnoncelli, M.G.B.; Vandenberghe, L.P.D.S.; Soccol, C.R. Microbiological, Biochemical, and Functional Aspects of Sugary Kefir Fermentation—A Review. Food Microbiol. 2017, 66, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Zanirati, D.F.; Abatemarco, M.J.; de Cicco Sandes, S.H.; Nicoli, J.R.; Nunes, Á.C.; Neumann, E. Selection of Lactic Acid Bacteria from Brazilian Kefir Grains for Potential Use as Starter or Probiotic Cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Bengoa, A.A.; Iraporda, C.; Garrote, G.L.; Abraham, A.G. Kefir Micro-Organisms: Their Role in Grain Assembly and Health Properties of Fermented Milk. J. Appl. Microbiol. 2019, 126, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Zhang, T.; Zhang, P.; He, X.; Sadiq, F.A.; Li, J.; Sang, Y.; Gao, J. The Complex World of Kefir: Structural Insights and Symbiotic Relationships. Compr. Rev. Food Sci. Food Saf. 2024, 23, e13364. [Google Scholar] [CrossRef] [PubMed]
- Blasche, S.; Kim, Y.; Mars, R.A.T.; Machado, D.; Maansson, M.; Kafkia, E.; Milanese, A.; Zeller, G.; Teusink, B.; Nielsen, J.; et al. Metabolic Cooperation and Spatiotemporal Niche Partitioning in a Kefir Microbial Community. Nat. Microbiol. 2021, 6, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Liu, B.; Jiang, T.; Liu, Y.; Chen, L. The Biofilm Hypothesis: The Formation Mechanism of Tibetan Kefir Grains. Int. J. Dairy Technol. 2018, 71, 44–50. [Google Scholar] [CrossRef]
- Nejati, F.; Junne, S.; Neubauer, P. A Big World in Small Grain: A Review of Natural Milk Kefir Starters. Microorganisms 2020, 8, 192. [Google Scholar] [CrossRef] [PubMed]
- Londero, A.; Hamet, M.F.; De Antoni, G.L.; Garrote, G.L.; Abraham, A.G. Kefir Grains as a Starter for Whey Fermentation at Different Temperatures: Chemical and Microbiological Characterisation. J. Dairy Res. 2012, 79, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Azizi, N.F.; Kumar, M.R.; Yeap, S.K.; Abdullah, J.O.; Khalid, M.; Omar, A.R.; Osman, M.A.; Mortadza, S.A.S.; Alitheen, N.B. Kefir and Its Biological Activities. Foods 2021, 10, 1210. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Jeong, D.; Kim, H.; Seo, K.H. Modern Perspectives on the Health Benefits of Kefir in next Generation Sequencing Era: Improvement of the Host Gut Microbiota. Crit. Rev. Food Sci. Nutr. 2019, 59, 1782–1793. [Google Scholar] [CrossRef] [PubMed]
- M’hir, S.; Ayed, L.; De Pasquale, I.; Fanizza, E.; Tlais, A.Z.A.; Comparelli, R.; Verni, M.; Latronico, R.; Gobbetti, M.; Di Cagno, R.; et al. Comparison of Milk Kefirs Obtained from Cow’s, Ewe’s and Goat’s Milk: Antioxidant Role of Microbial-Derived Exopolysaccharides. Antioxidants 2024, 13, 335. [Google Scholar] [CrossRef] [PubMed]
- Amorim, F.G.; Coitinho, L.B.; Dias, A.T.; Friques, A.G.F.; Monteiro, B.L.; de Rezende, L.C.D.; Pereira, T.d.M.C.; Campagnaro, B.P.; De Pauw, E.; Vasquez, E.C.; et al. Identification of New Bioactive Peptides from Kefir Milk through Proteopeptidomics: Bioprospection of Antihypertensive Molecules. Food Chem. 2019, 282, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, J.; Du, L.; Chen, J.; Zheng, Q.; Li, P.; Du, B.; Fang, X.; Liao, Z. Kefir Microbiota and Metabolites Stimulate Intestinal Mucosal Immunity and Its Early Development. Crit. Rev. Food Sci. Nutr. 2024, 64, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequencing-Based Analysis of the Bacterial and Fungal Composition of Kefir Grains and Milks from Multiple Sources. PLoS ONE 2013, 8, e69371. [Google Scholar] [CrossRef] [PubMed]
- González-Orozco, B.D.; García-Cano, I.; Escobar-Zepeda, A.; Jiménez-Flores, R.; Álvarez, V.B. Metagenomic Analysis and Antibacterial Activity of Kefir Microorganisms. J. Food Sci. 2023, 88, 2933–2949. [Google Scholar] [CrossRef] [PubMed]
- Walsh, A.M.; Crispie, F.; Kilcawley, K.; O’Sullivan, O.; O’Sullivan, M.G.; Claesson, M.J.; Cotter, P.D. Microbial Succession and Flavor Production in the Fermented Dairy Beverage Kefir. mSystems 2016, 1, e00052-16. [Google Scholar] [CrossRef]
- Villanoeva, C.; Rios, D.; Alvarenga, R.; Acurcio, L.; Sandes, S.; Nunes, A.; Nicoli, J.; Neumann, E. Functionally Active Microbiome and Physicochemical Properties of Milk and Sugary Water Kefir from Brazil. Austin Food Sci. 2021, 6, 1–8. [Google Scholar] [CrossRef]
- McGovern, C.J.; González-Orozco, B.D.; Jiménez-Flores, R. Evaluation of Kefir Grain Microbiota, Grain Viability, and Kefir Bioactivity from Fermenting Dairy Processing By-Products. J. Dairy Sci. 2024, 107, 4259–4276. [Google Scholar] [CrossRef] [PubMed]
- Satir, G.; Guzel-Seydim, Z.B. How Kefir Fermentation Can Affect Product Composition? Small Rumin. Res. 2016, 134, 1–7. [Google Scholar] [CrossRef]
- Yegin, Z.; Yurt, M.N.Z.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Ucak, S.; Ozalp, V.C.; Sudagidan, M. Determination of Bacterial Community Structure of Turkish Kefir Beverages via Metagenomic Approach. Int. Dairy J. 2022, 129, 105337. [Google Scholar] [CrossRef]
- Walsh, L.H.; Coakley, M.; Walsh, A.M.; Crispie, F.; O’Toole, P.W.; Cotter, P.D. Analysis of the Milk Kefir Pan-Metagenome Reveals Four Community Types, Core Species, and Associated Metabolic Pathways. iScience 2023, 26, 108004. [Google Scholar] [CrossRef] [PubMed]
- Gentry, B.; Cazón, P.; O’Brien, K. A Comprehensive Review of the Production, Beneficial Properties, and Applications of Kefiran, the Kefir Grain Exopolysaccharide. Int. Dairy J. 2023, 144, 105691. [Google Scholar] [CrossRef]
- Simonelli, N.; Gagliarini, N.; Medrano, M.; Piermaria, J.; Abraham, A. Kefiran. In Polysaccharides of Microbial Origin; Oliveira, J., Radhouani, H., Reis, R., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 1–12. ISBN 9783662467640. [Google Scholar]
- Rimada, P.S.; Abraham, A.G. Polysaccharide Production by Kefir Grains during Whey Fermentation. J. Dairy Res. 2001, 68, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Rimada, P.S.; Abraham, A.G. Comparative Study of Different Methodologies to Determine the Exopolysaccharide Produced by Kefir Grains in Milk and Whey. Lait 2003, 83, 79–87. [Google Scholar] [CrossRef]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Characteristics of Kefir Prepared with Different Grain:Milk Ratios. J. Dairy Res. 1998, 65, 149–154. [Google Scholar] [CrossRef]
- Bourrie, B.C.T.; Ju, T.; Fouhse, J.M.; Forgie, A.J.; Sergi, C.; Cotter, P.D.; Willing, B.P. Kefir Microbial Composition Is a Deciding Factor in the Physiological Impact of Kefir in a Mouse Model of Obesity. Br. J. Nutr. 2021, 125, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef]
- Rios, D.L.; da Silva, P.C.L.; Moura, C.S.S.; Villanoeva, C.N.B.C.; da Rocha Fernandes, G.; Bengoa, A.A.; Garrote, G.L.; Abraham, A.G.; Nicoli, J.R.; Neumann, E.; et al. Comparative Metatranscriptome Analysis of Brazilian Milk and Water Kefir Beverages. Int. Microbiol. 2023, 27, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Vasapolli, R.; Schütte, K.; Schulz, C.; Vital, M.; Schomburg, D.; Pieper, D.H.; Vilchez-Vargas, R.; Malfertheiner, P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology 2019, 157, 1081–1092.e3. [Google Scholar] [CrossRef] [PubMed]
- Garrote, G.L.; Abraham, A.G.; De Antoni, G.L. Inhibitory Power of Kefir: The Role of Organic Acids. J. Food Prot. 2000, 63, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Hamet, M.F.; Piermaria, J.A.; Abraham, A.G. Selection of EPS-Producing Lactobacillus Strains Isolated from Kefir Grains and Rheological Characterization of the Fermented Milks. LWT Food Sci. Technol. 2015, 63, 129–135. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High Speed BLASTN: An Accelerated MegaBLAST Search Tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Tappu, R.; Bazinet, A.L.; Xie, C.; Cummings, M.P.; Nieselt, K.; Williams, R. Fast and Simple Protein-Alignment-Guided Assembly of Orthologous Gene Families from Microbiome Sequencing Reads. Microbiome 2017, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Lomsadze, A.; Borodovsky, M. Identification of Protein Coding Regions in RNA Transcripts. Nucleic Acids Res. 2015, 43, e78. [Google Scholar] [CrossRef] [PubMed]
- Mai, H.; Zhang, Y.; Li, D.; Leung, H.C.-M.; Luo, R.; Wong, C.-K.; Ting, H.-F.; Lam, T.-W. AC-DIAMOND v1: Accelerating Large-Scale DNA-Protein Alignment. Bioinformatics 2018, 34, 3744–3746. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, G.R.; Barbosa, D.V.C.; Prosdocimi, F.; Pena, I.A.; Santana-Santos, L.; Coelho Junior, O.; Barbosa-Silva, A.; Velloso, H.M.; Mudado, M.A.; Natale, D.A.; et al. A Procedure to Recruit Members to Enlarge Protein Family Databases--the Building of UECOG (UniRef-Enriched COG Database) as a Model. Genet. Mol. Res. 2008, 7, 910–924. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Low, J.Z.B.; Khang, T.F.; Tammi, M.T. CORNAS: Coverage-Dependent RNA-Seq Analysis of Gene Expression Data without Biological Replicates. BMC Bioinform. 2017, 18, 575. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Rimada, P.S.; Abraham, A.G. Kefiran Improves Rheological Properties of Glucono-δ-Lactone Induced Skim Milk Gels. Int. Dairy J. 2006, 16, 33–39. [Google Scholar] [CrossRef]
- Barros, S.É.D.L.; Rocha, C.D.S.; De Moura, M.S.B.; Barcelos, M.P.; Da Silva, C.H.T.D.P.; Hage-Melim, L.I.D.S. Potential Beneficial Effects of Kefir and Its Postbiotic, Kefiran, on Child Food Allergy. Food Funct. 2021, 12, 3770–3786. [Google Scholar] [CrossRef]
- Salvetti, E.; O’Toole, P.W. The Genomic Basis of Lactobacilli as Health-Promoting Organisms. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Valasaki, K.; Staikou, A.; Theodorou, L.G.; Charamopoulou, V.; Zacharaki, P.; Papamichael, E.M. Purification and Kinetics of Two Novel Thermophilic Extracellular Proteases from Lactobacillus Helveticus, from Kefir with Possible Biotechnological Interest. Bioresour. Technol. 2008, 99, 5804–5813. [Google Scholar] [CrossRef]
- Cheirsilp, B.; Suksawang, S.; Yeesang, J.; Boonsawang, P. Co-Production of Functional Exopolysaccharides and Lactic Acid by Lactobacillus Kefiranofaciens Originated from Fermented Milk, Kefir. J. Food Sci. Technol. 2018, 55, 331–340. [Google Scholar] [CrossRef]

| MKAA1 | MKAA2 | |
|---|---|---|
| LAB (log CFU/mL) | 9.32 ± 0.13 | 9.17 ± 0.08 |
| Yeast (log CFU/mL) | 6.24 ± 0.62 | 6.33 ± 0.31 |
| pH | 4.28 ± 0.02 a | 4.07 ± 0.02 b |
| Lactic acid (mM) | 86.96 ± 6.6 | 106.67 ± 6.1 |
| Acetic acid (mM) | 30.78 ± 0.9 a | 57.08 ± 5.8 b |
| Viscosity at 300 s−1 (mPa.s) | 44.05 ± 6.6 a | 26.83 ± 0.47 b |
| Library * | Raw Reads | HS-BLASTN Reads | TransDecoder Reads | KO Protein Reads | Annotated Reads | % Total Reads |
|---|---|---|---|---|---|---|
| MKAA1_y | 1,345,933 | 1,201,797 | 771,559 | 568,621 | 1,276,753 | 94.86% |
| MKAA2_y | 1,046,458 | 800,109 | 739,280 | 572,576 | 950,221 | 90.80% |
| MKAA1_b | 4,113,272 | 4,028,464 | 3,060,355 | 2,648,774 | 4,055,211 | 98.59% |
| MKAA2_b | 2,530,065 | 2,386,120 | 1,757,839 | 1,518,057 | 2,449,472 | 96.81% |
| Library * | Trinity Contigs | TransDecoder mRNA Contigs | % mRNA | KO protein Contigs | % KO |
|---|---|---|---|---|---|
| MKAA1_y | 57,837 | 24,010 | 41.51% | 17,082 | 29.53% |
| MKAA2_y | 63,859 | 30,170 | 47.24% | 22,199 | 34.76% |
| MKAA1_b | 41,450 | 18,139 | 43.76% | 14,057 | 33.91% |
| MKAA2_b | 50,018 | 20,243 | 40.47% | 15,696 | 31.38% |
| Lactic Acid Bacteria Species | MKAA1 | MKAA2 | Both | Shared | MKAA1-Specific | MKAA2-Specific |
|---|---|---|---|---|---|---|
| Lactococcus lactis | 1068 (90.6%) | 1057 (89.7%) | 1179 | 946 (80.2%) | 122 (10.3%) | 111 (9.4%) |
| Leuconostoc mesenteroides | 881 (90.5%) | 866 (88.9%) | 974 | 773 (79.4%) | 108 (11.1%) | 93 (9.5%) |
| Lactobacillus kefiranofaciens | 941 (93.6%) | 903 (89.9%) | 1005 | 839 (83.5%) | 102 (10.1%) | 64 (6.4%) |
| Lactobacillus helveticus | 900 (97.5%) | 547 (59.3%) | 923 | 524 (56.8%) | 376 (40.7%) | 23 (2.5%) |
| Lactobacillus gallinarum | 791 (98.4%) | 404 (50.2%) | 804 | 391 (48.6%) | 400 (49.8%) | 13 (1.6%) |
| Overall number of KO | 1454 (93.5%) | 1428 (91.8%) | 1555 | 1327 (85.3%) | 127 (8.2%) | 101 (6.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lisboa Rios, D.; Bengoa, A.A.; Costa Lima da Silva, P.; Moura, C.S.S.; Garrote, G.L.; Abraham, A.G.; da Rocha Fernandes, G.; Nicoli, J.R.; Neumann, E.; Cantini Nunes, Á. Metatranscriptomic Analysis of Argentinian Kefirs Varying in Apparent Viscosity. Appl. Microbiol. 2024, 4, 1150-1164. https://doi.org/10.3390/applmicrobiol4030078
Lisboa Rios D, Bengoa AA, Costa Lima da Silva P, Moura CSS, Garrote GL, Abraham AG, da Rocha Fernandes G, Nicoli JR, Neumann E, Cantini Nunes Á. Metatranscriptomic Analysis of Argentinian Kefirs Varying in Apparent Viscosity. Applied Microbiology. 2024; 4(3):1150-1164. https://doi.org/10.3390/applmicrobiol4030078
Chicago/Turabian StyleLisboa Rios, Diego, Ana Agustina Bengoa, Patrícia Costa Lima da Silva, César Silva Santana Moura, Graciela Liliana Garrote, Analía Graciela Abraham, Gabriel da Rocha Fernandes, Jacques Robert Nicoli, Elisabeth Neumann, and Álvaro Cantini Nunes. 2024. "Metatranscriptomic Analysis of Argentinian Kefirs Varying in Apparent Viscosity" Applied Microbiology 4, no. 3: 1150-1164. https://doi.org/10.3390/applmicrobiol4030078
APA StyleLisboa Rios, D., Bengoa, A. A., Costa Lima da Silva, P., Moura, C. S. S., Garrote, G. L., Abraham, A. G., da Rocha Fernandes, G., Nicoli, J. R., Neumann, E., & Cantini Nunes, Á. (2024). Metatranscriptomic Analysis of Argentinian Kefirs Varying in Apparent Viscosity. Applied Microbiology, 4(3), 1150-1164. https://doi.org/10.3390/applmicrobiol4030078






