Escherichia coli Reporter Strains Allow for the In Vivo Evaluation of Recombinant Elongation Factor Protein (EF-P)
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and DNA Primers
2.2. Construction of Plasmids and Recombinant Strains
2.3. Growth Measurements
2.4. Biochemical Measurements
3. Results and Discussion
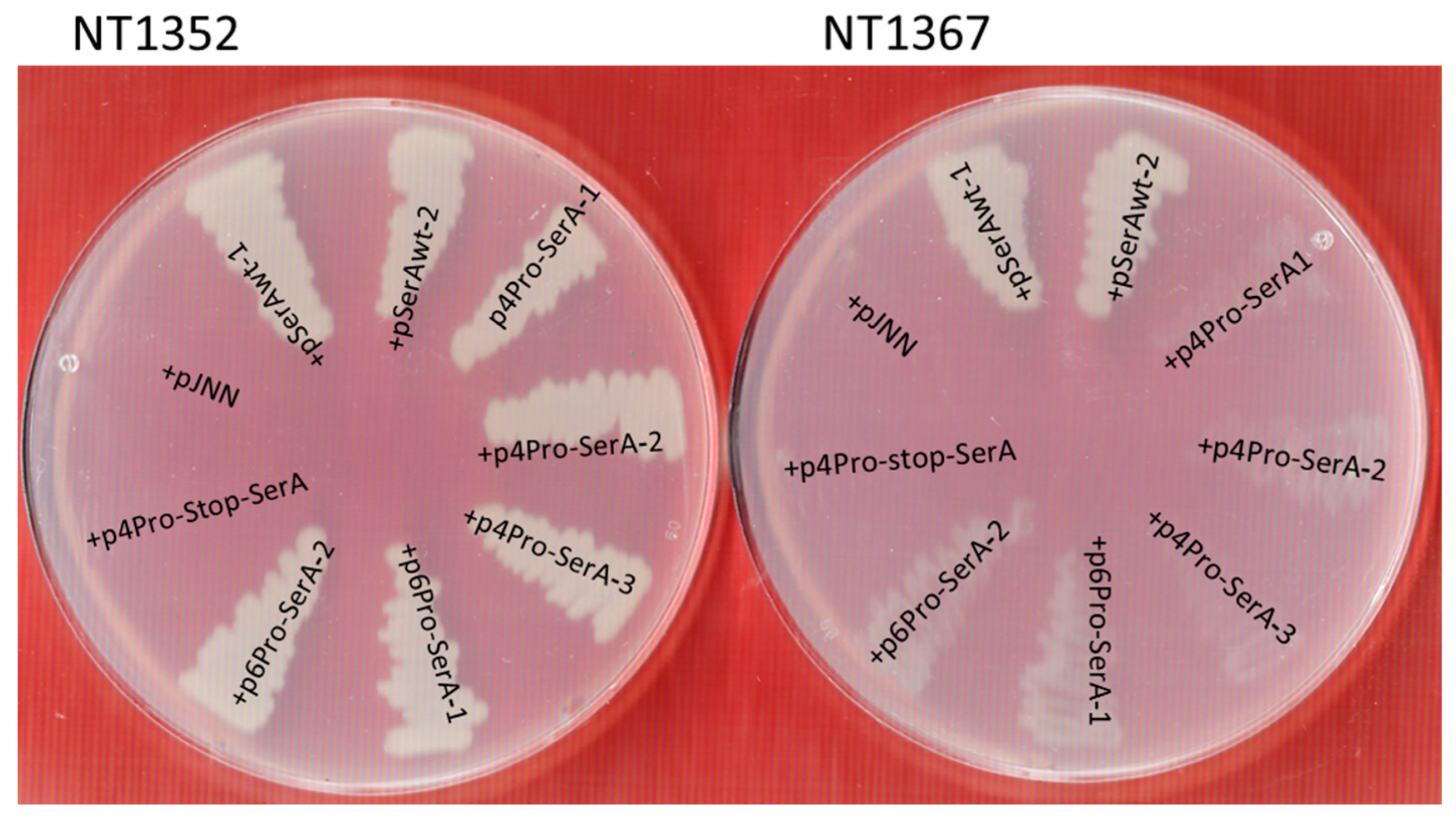
3.1. Construction of E. coli Reporter Strains Which Lack Their Own EF-P-System
3.2. Synthetic Reporter Strains Which Show a Growth Defect in Mineral Media
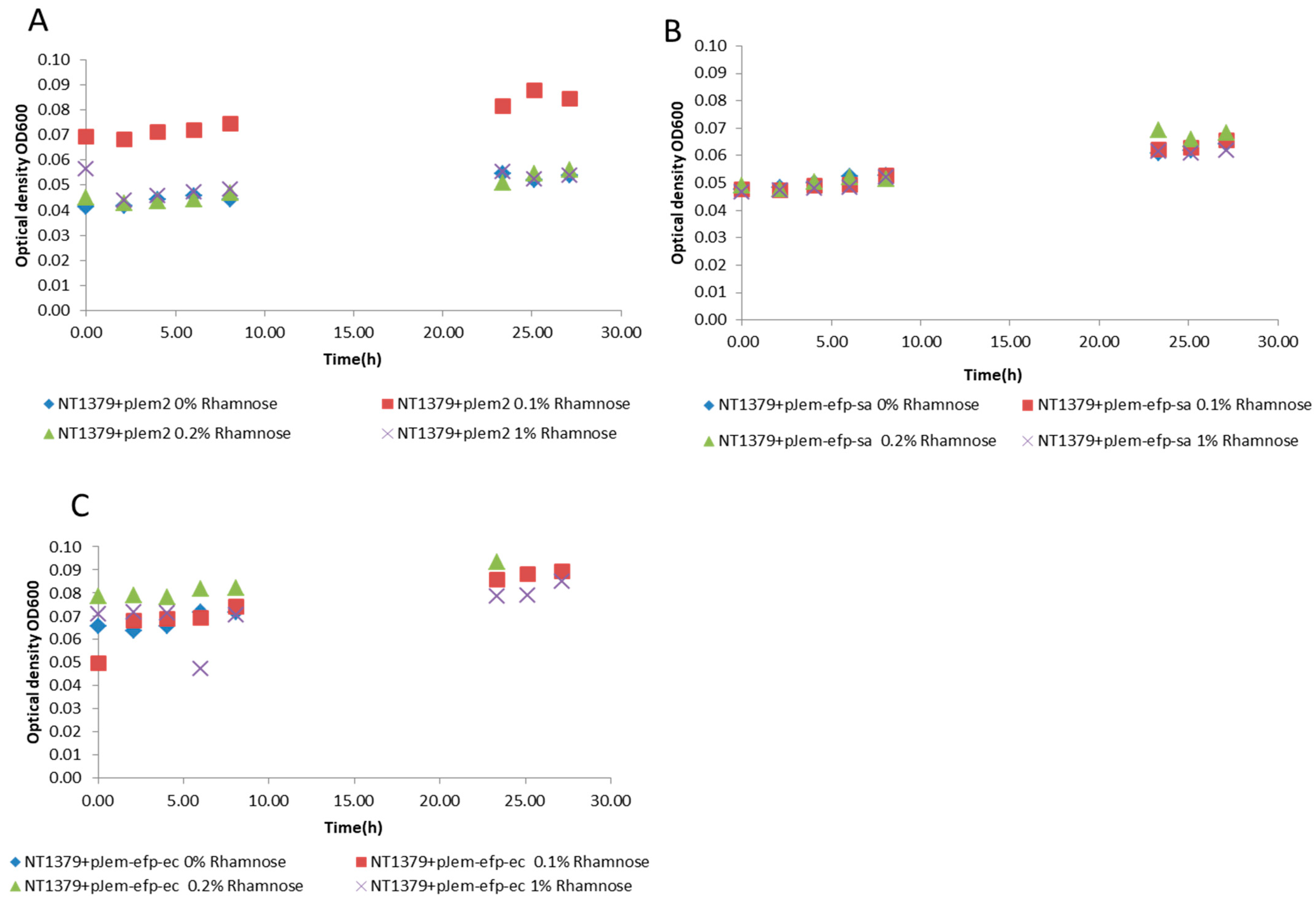
3.3. Plasmid-Based Expression of Efp Genes
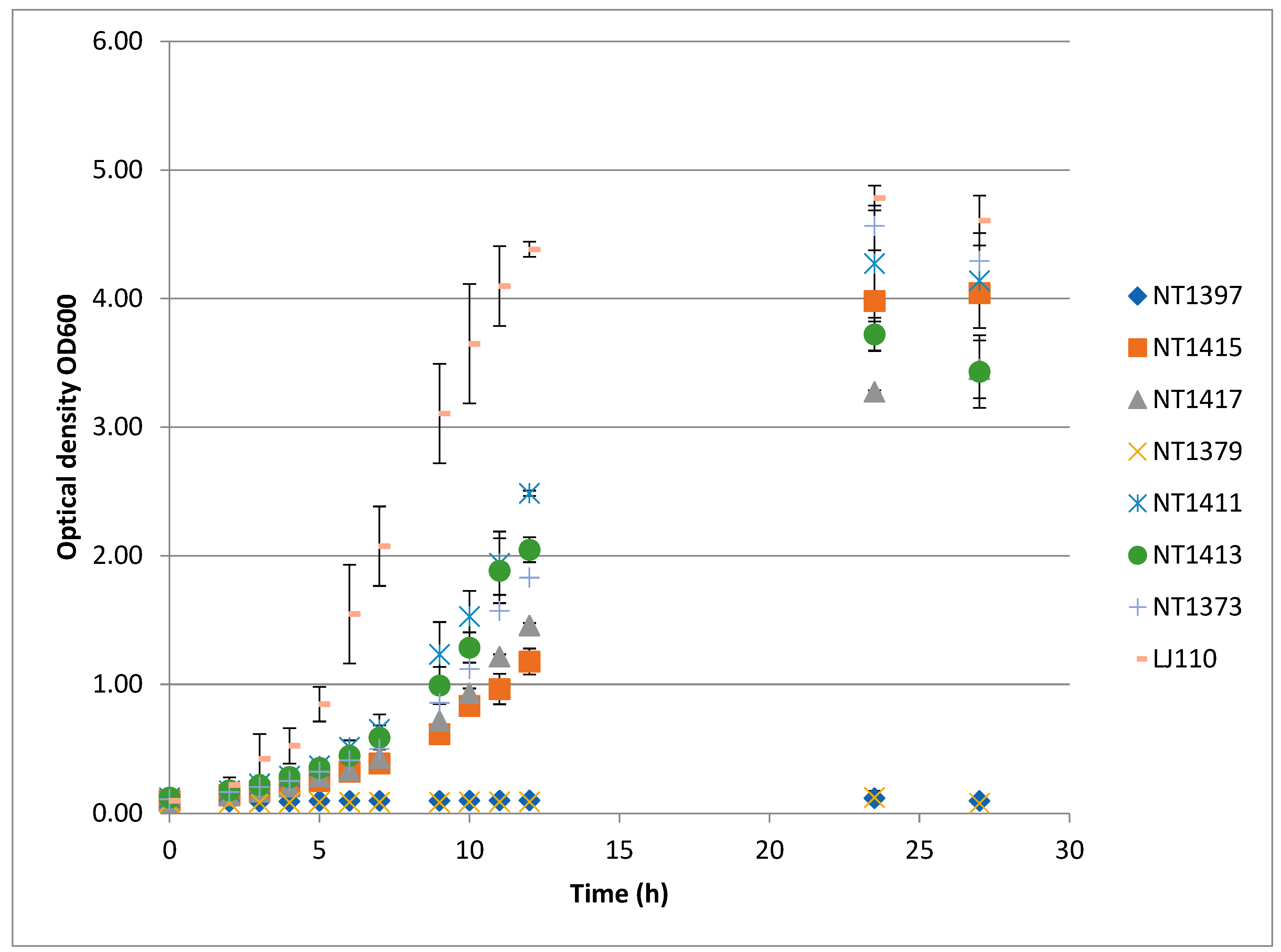
3.4. Complementation of Growth Defects by Chromosomally Integrated Recombinant Efp Genes
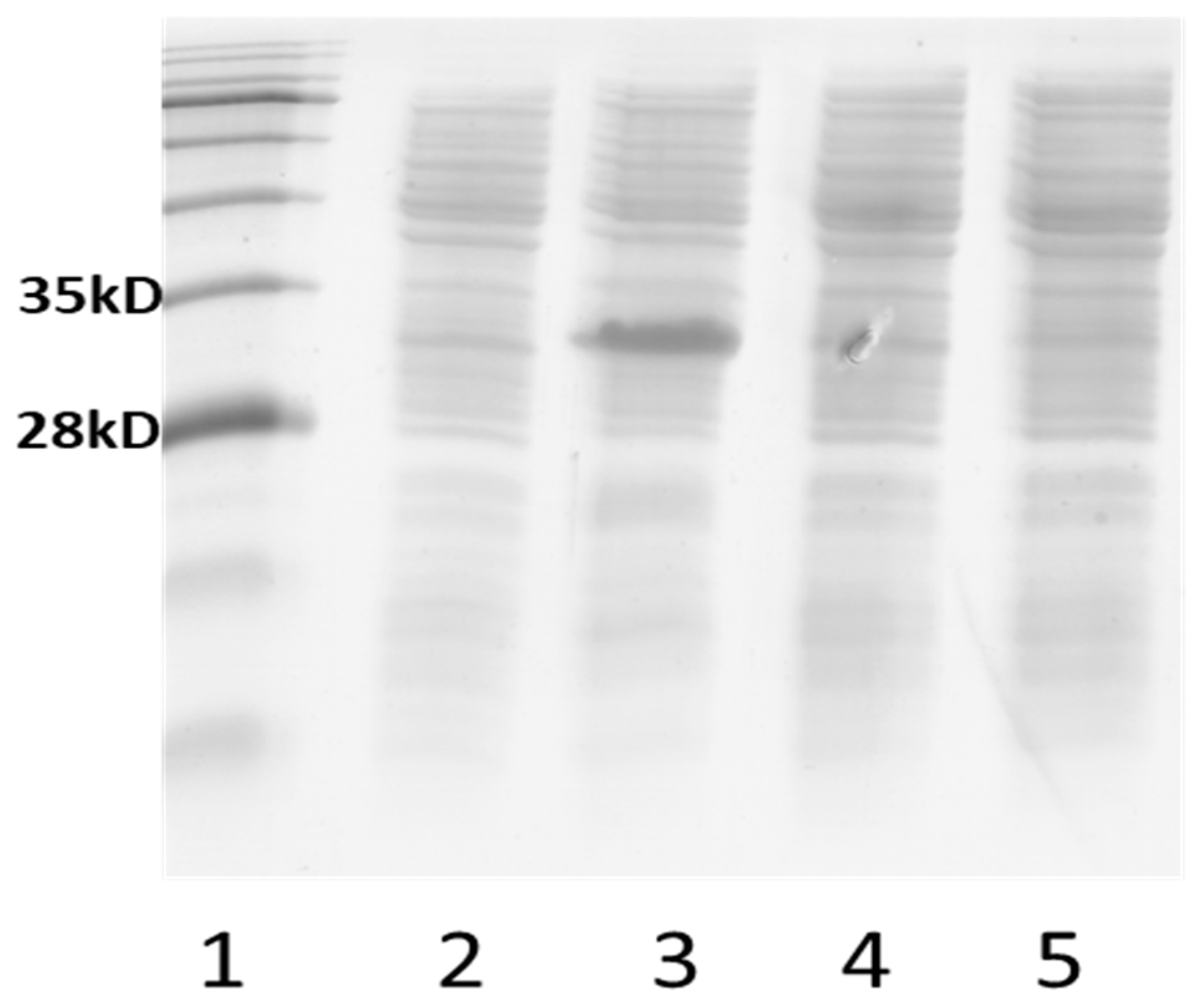
3.5. Transcript Analyses of Recombinant Strains
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ude, S.; Lassak, J.; Starosta, A.L.; Kraxenberger, T.; Wilson, D.N.; Jung, K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 2013, 339, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Peil, L.; Starosta, A.L.; Lassak, J.; Atkinson, G.C.; Virumae, K.; Spitzer, M.; Tenson, T.; Jung, K.; Remme, J.; Wilson, D.N. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc. Natl. Acad. Sci. USA 2013, 110, 15265–15270. [Google Scholar] [CrossRef] [PubMed]
- Doerfel, L.K.; Wohlgemuth, I.; Kothe, C.; Peske, F.; Urlaub, H.; Rodnina, M.V. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 2013, 339, 85–88. [Google Scholar] [CrossRef]
- Huter, P.; Arenz, S.; Bock, L.V.; Graf, M.; Frister, J.O.; Heuer, A.; Peil, L.; Starosta, A.L.; Wohlgemuth, I.; Peske, F.; et al. Structural basis for polyproline-mediated ribosome stalling and rescue by the translational elongation factor EF-P. Mol. Cell 2017, 68, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Bailly, M.; de Crécy-Lagard, V. Predicting the pathway involved in post-translational modification of elongation factor P in a subset of bacterial species. Biol. Direct. 2010, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Kuroshu, R.; Zanelli, C.F.; Valentini, S.R. eIF5A and EF-P: Two unique translation factors are now traveling the same road. Wiley Interdiscip. Rev. RNA. 2014, 5, 209–222. [Google Scholar] [CrossRef]
- Rodnina, M.V. Translation in Prokaryotes. Cold Spring Harb. Perspect. Biol. 2018, 10, a032664. [Google Scholar] [CrossRef]
- Hummels, K.R.; Kearns, D.B. Translation elongation factor P (EF-P). FEMS Microbiol. Rev. 2020, 44, 208–218. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, S.; Fu, X.; Dedkova, L.M.; Hecht, S.M. Enhancement of N-Methyl amino acid incorporation into proteins and peptides using modified bacterial ribosomes and elongation factor P. ACS Chem. Biol. 2024, 19, 1330–1338. [Google Scholar] [CrossRef]
- Hanawa-Suetsugu, K.; Sekine, S.I.; Sakai, H.; Hori-Takemoto, C.; Terada, T.; Unzai, S.; Tame, J.R.; Kuramitsu, S.; Shirouzu, M.; Yokoyama, S. Crystal structure of elongation factor P from Thermus thermophilus HB8. Proc. Natl. Acad. Sci. USA 2004, 101, 9595–9600. [Google Scholar] [CrossRef]
- Choi, S.; Choe, J. Crystal structure of elongation factor P from Pseudomonas aeruginosa at 1.75 Å resolution. Proteins 2011, 79, 1688–1693. [Google Scholar] [CrossRef] [PubMed]
- Blaha, G.; Stanley, R.E.; Steitz, T.A. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science 2009, 325, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Rajkovic, A.; Ibba, M. Elongation factor P and the control of translation elongation. Annu. Rev. Microbiol. 2017, 71, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Lassak, J.; Keilhauer, E.; Fürst, M.; Wuichet, K.; Gödeke, J.; Starosta, A.L.; Chen, J.-M.; Sogaard-Andersen, L.; Rohr, J.; Wilson, D.N.; et al. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat. Chem. Biol. 2015, 11, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Lassak, J.; Sieber, A.; Hellwig, M. Exceptionally versatile take II: Post-translational modifications of lysine and their impact on bacterial physiology. Biol. Chem. 2022, 403, 819–858. [Google Scholar] [CrossRef]
- Rajkovic, A.; Hummels, K.R.; Witzky, A.; Erickson, S.; Gafken, P.R.; Whitelegge, J.P.; Faull, K.F.; Kearns, D.B.; Ibba, M. Translation control of swarming proficiency in Bacillus subtilis by 5-Amino-pentanolylated Elongation Factor P. J. Biol. Chem. 2016, 291, 10976–10985. [Google Scholar] [CrossRef]
- Witzky, A.; Hummels, K.R.; Tollerson, R.; Rajkovic, A.; Jones, L.A.; Kearns, D.B.; Ibba, M. EF-P posttranslational modification has variable impact on polyproline translation in Bacillus subtilis. mBio 2018, 9, e00306-1. [Google Scholar] [CrossRef]
- Golubev, A.; Negroni, L.; Krasnovid, F.; Validov, S.; Yusupova, G.; Yusupov, M.; Usachev, K. Posttranslational modification of Elongation Factor P from Staphylococcus aureus. FEBS Open Bio 2020, 10, 1342–1347. [Google Scholar] [CrossRef]
- Pinheiro, B.; Scheidler, C.M.; Kielkowski, P.; Schmid, M.; Forné, I.; Ye, S.; Reiling, N.; Takano, E.; Imhof, A.; Sieber, S.A.; et al. Structure and Function of an Elongation Factor P Subfamily in Actinobacteria. Cell Rep. 2020, 30, 4332–4342.e5. [Google Scholar] [CrossRef]
- Ohashi, Y.; Inaoka, T.; Kasai, K.; Ito, Y.; Okamoto, S.; Satsu, H.; Tozawa, Y.; Kawamura, F.; Ochi, K. Expression profiling of translation-associated genes in sporulating Bacillus subtilis and consequence of sporulation by gene inactivation. Biosci. Biotechnol. Biochem. 2003, 67, 2245–2253. [Google Scholar] [CrossRef]
- Feaga, H.A.; Hong, H.-R.; Prince, C.R.; Rankin, A.; Buskirk, A.R.; Dworkin, J. Elongation factor P is important for sporulation initiation. J. Bacteriol. 2023, 205, e00370-22. [Google Scholar] [CrossRef] [PubMed]
- Balibar, C.J.; Iwanowicz, D.; Dean, C.R. Elongation factor P is dispensable in Escherichia coli and Pseudomonas aeruginosa. Curr. Microbiol. 2013, 67, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Dekany, K.; Adams, S.L.; Ganoza, M.C. The gene encoding the elongation factor P protein is essential for viability and is required for protein synthesis. J. Biol. Chem. 1997, 272, 32254–32259. [Google Scholar] [CrossRef]
- Krafczyk, R.; Macošek, J.; Jagtap, P.K.A.; Gast, D.; Wunder, S.; Mitra, P.; Jha, A.K.; Rohr, J.; Hoffmann-Röder, A.; Jung KHennig, J.; et al. Structural Basis for EarP-Mediated Arginine Glycosylation of Translation Elongation Factor EF-P. mBio 2017, 8, e01412-17. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.R.; Allen, A.G.; Owen, P.J.; Shalom, G.; Stone, K.; Harrison, M.; Burgis, T.A.; Lockyer, M.; Garcia-Lara, J.; Foster, S.J.; et al. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genom. 2009, 10, 291. [Google Scholar] [CrossRef]
- Rajkovic, A.; Erickson, S.; Witzky, A.; Branson, O.E.; Seo, J.; Gafken, P.R.; Frietas, M.A.; Whitelegge, J.P.; Fauli, K.F.; Navarre, W.; et al. Cyclic rhamnosylated Elongation Factor P establishes antibiotic resistance in Pseudomonas aeruginosa. mBio 2015, 6, e00823. [Google Scholar] [CrossRef]
- Stauffer, G.V. Regulation of serine, glycine and one-carbon biosynthesis. EcoSalPlus 2013, 1. [Google Scholar] [CrossRef]
- Zeppenfeld, T.; Larisch, C.; Lengeler, J.W.; Jahreis, K. Glucose transporter mutants of Escherichia coli K-12 with changes in substrate recognition of IICB(Glc) and induction behavior of the ptsG gene. J. Bacteriol. 2000, 182, 4443–4452. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef]
- Jeske, M.; Altenbuchner, J. The Escherichia coli rhamnose promoter rhaP(BAD) is in Pseudomonas putida KT2440 independent of Crp-cAMP activation. Appl. Microbiol. Biotechnol. 2010, 85, 1923–1933. [Google Scholar] [CrossRef]
- Rodrigues, A.L.; Trachtmann, N.; Becker, J.; Lohanatha, A.F.; Blotenberg, J.; Bolten, C.J.; Korneli, C.; de Souza Lima, A.O.; Porto, L.M.; Sprenger, G.A.; et al. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 2013, 20, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. (Eds.) Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 978-087969577-4. [Google Scholar]
- Albermann, C.; Trachtmann, N.; Sprenger, G.A. A simple and reliable method to conduct and monitor expression cassette integration into the Escherichia coli chromosome. Biotechnol. J. 2010, 5, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Woodcock, D.M.; Crowther, P.J.; Doherty, J.; Jefferson, S.; DeCruz, E.; Noyer-Weidner, M.; Smith, S.S.; Michael, M.Z.; Graham, M.W. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989, 17, 3469–3478. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Lerner, S.A.; Lin, E.C.C. Replacement of a phosphoenolpyruvate-dependent phosphotransferase by a nicotinamide adenine dinucleotide-linked dehydrogenase for the utilization of mannitol. J. Bacteriol. 1967, 93, 642–648. [Google Scholar] [CrossRef]

| Strains | ||
|---|---|---|
| Name | Relevant Genotype | Reference |
| LJ110 | E. coli K-12 W3110, wild-type, prototroph | [28] |
| NT1352 | LJ110 ΔserA | This work |
| NT1367 | LJ110 ΔserA Δefp | This work |
| NT1368 | LJ110 ΔserA fuc::PserA-serA Δefp | This work |
| NT1373 | LJ110 ΔserA fuc::PserA 6Pro-serA | This work |
| NT1379 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA | This work |
| NT1397 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA ΔepmA | This work |
| NT 1411 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA xyl::Ptac-efp-Ec * | This work |
| NT 1413 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA xyl::Ptac-efp-Sa ** | This work |
| NT 1415 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA ΔepmA xyl::Ptac-efp-Ec * | This work |
| NT 1417 | LJ110 Δefp ΔserA fuc::PserA 6Pro-serA ΔepmA xyl::Ptac-efp-Sa ** | This work |
| NT1433 | LJ110 ΔepmA | This work |
| NT1435 | LJ110 Δefp | This work |
| NT1437 | LJ110 Δefp ΔepmA | This work |
| Plasmids | ||
| pTarget F | pMB1 aadA sgRNA | [29] |
| pCas | repA101(Ts)kan Pcas-cas9 ParaB-λRed lacIq Plac-sgRNA-pMB1 | [29] |
| pTarget–cat | catR-cassette into the BclI site of the pTargetF plasmid with disruption of Smr gene | This work |
| pTarget-catsg-fuc | pMB1 aadA sgRNA-fuc | This work |
| pTarget-catsg-serA | pMB1 aadA sgRNA-serA | This work |
| pTarget-catsg-efp | pMB1 aadA sgRNA-efp | This work |
| pTarget-catsg-epmA | pMB1 aadA sgRNA-serA | This work |
| pJem2 | rhaR rhaS rhaP BAD –T7le-eGFP , mob, kan | [30] |
| pJNNmod | ColE1 Ptac,lacIq,bla | [31] |
| pSerA | ColE, lacIq, PserA-serA | This work |
| p6Pro-serA | ColE1, lacIq, PserA-6Pro-serA | This work |
| p4Pro-serA | ColE1, lacIq, PserA-4Pro-serA | This work |
| p6Pro-stop-serA | ColE1, lacIq, PserA-4Pro-UAA-serA | This work |
| pJNN-efp-Ec * | ColE1 lacIqPtac-efp (E. coli) | This work |
| pJNN-efp-Sa ** | ColE1 lacIqPtac-efp ** | This work |
| pJem2-efp-Ec * | rhaR rhaS Prha –efp-Ec *, mob, kan | This work |
| pJem2-efp-Sa ** | rhaR rhaS Prha –efp-Sa *, mob, kan | This work |
| Name | DNA Sequence |
|---|---|
| Targ-univ-rev | actagtattatacctaggactgagctagc |
| sgRNA-serA | ggcattctggctgaatcgctgttttagagctagaaatagcaagttaaaataaggctag |
| sgRNA-efp | aaaccggctaccctgtctacgttttagagctagaaatagcaagttaaaataaggctag |
| sgRNA-epmA | gagacacgtttcgttggcccgttttagagctagaaatagcaagttaaaataaggctag |
| sgRNA-fuc | gacgaccgtcaataaccggggttttagagctagaaatagcaagttaaaataaggctag |
| sgRNA-xyl | gcccaattcgctattccagcgttttagagctagaaatagcaagttaaaataaggctag |
| 3′serA-SphI | ttttgcatgcttagtacagcagacgggcgcgaatg |
| 5′PserA-serA-SphI | ttttgcatgcctcttcattaaatttggtgacatgtgtcacg |
| serA-inv-rev | ttacccaatcctgtcttttgaaatgttgtg |
| serA-4Pro | atgccaccaccaccagcaaaggtatcgctggagaaagacaag |
| serA-6Pro | atgccaccaccaccaccaccagcaaaggtatcgctggagaaagacaag |
| QC-Stop-SerA-I | caggattgggtaaatgtaaccaccaccaccagc |
| QC-Stop-SerA-II | tggtggtggtggttacatttacccaatcctgtcttttg |
| efp-Ec-5-NdeI | ttttcatatggcaacgtactatagcaacgattttc |
| efp-Ec-3′HindIII | ttttaagcttacttcacgcgagagacgtattc |
| efp-Sa-3′-BamHI | ttttggatccttatcctcttgaaatgtagcttccatcacc |
| efp-Sa-5′-NdeI | ttttcatatgggcatttcggttaatgattttaaaacagg |
| L5′del-SerA | cgcgtcagctggtgaaactgggcg |
| L3′del-SerA-BamHI | ttttggatccagagcaatcgacaattgcctgg |
| R5′del-SerA-BglII | ttttagatctcccgtctgctgtactaattcccc |
| R3′del-SerA | gggtaagggaggattgctcctccc |
| L5′-del-efp | gcgcgatgacaaactcatcttgcg |
| L3′-del-efp ′-BamHI | ttttggatcctctaacatgattttaagaccagcacg |
| R5′-del-efp BglII | ttttagatctatgcggttgtggtgcggcctg |
| R3′-del-efp | tgctgcgccagaaatcgcgttaccg |
| L5′-del-epmA-L-5 | tatcccacagccacgtacttcaggg |
| L3′-del-epmA-BamHI | ttttggatcctgacaagggcacgaagtctactcgc |
| R5′-del-epmA-BglII | ttttagatctactgaattaacagcgaagaatggcgtg |
| R3′-del-epmA | gcagctcccatttcagccatcattaagg |
| FucP-serA-int-5′ | tgctgtgctcactgttttttctttgggcggtagccaataaccttaacgacatgccccagcaggcgaaaatcctg |
| FucI-int-3′ | ggcgagagtgataaagtctgcgccaacgtggccgatggtcagaacccccagggttattgtctcatgagcg |
| xylA-int-5′ | gacgaactggtgttgggtaagcgtatggaagagcacttgcgttttgccgcctgctcaaggcgcactcccgttctgg |
| xylB-int-3′ | attaaagctgggacattgctcaggccggttaatttcgcggcccaatccagacaccagggttattgtctcatgagcg |
| RT-qPCR-serA 5′ | caagattaagtttctgctggtagaaggcg |
| RT-qPCR-serA 3′ | tagcgaccagtttttctgcggcgttg |
| RT-qPCR-ftsZ-5′ | tgcatttgcttccgacaacg |
| RT-qPCR-ftsZ-3′ | acgtttgtccatgccgatac |
| µ h−1 | fOD | |
|---|---|---|
| LJ110 | 0.58 +/− 0.01 | 7.14 +/− 0.51 |
| NT1433 | 0.41 +/− 0.03 | 4.87 +/− 0.28 |
| NT1435 | 0.5 +/− 0.03 | 5.27 +/− 0.16 |
| NT1437 | +/− 0.04 | 4.87 +/− 0.04 |
| µ h−1 | fOD | |
|---|---|---|
| LJ110 | 0.52 +/− 0.03 | 4.6 +/− 0.19 |
| NT1373 | 0.24 +/− 0.01 | 4.29 +/− 0.42 |
| NT1397 | n.a. | 0.09 +/−0.001 |
| NT1415 | 0.21 +/− 0.02 | 4.04 +/− 0.05 |
| NT1417 | 0.25 +/− 0.01 | 3.45 +/− 0.22 |
| NT1379 | n.a. | 0.08 +/− 0.01 |
| NT1411 | 0.26 +/− 0.001 | 4.14 +/− 0.36 |
| NT1413 | 0.29 +/− 0.02 | 3.43 +/− 0.28 |
| RQ | |
|---|---|
| LJ110 | 1.76 +/− 0.13 |
| NT1373 | 1.11 +/− 0.07 |
| NT1411 | 1.12 +/− 0.04 |
| NT1413 | 1.10 +/− 0.03 |
| NT1415 | 0.92 +/− 0.03 |
| NT1417 | 1.22 +/− 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trachtmann, N.; Bikmullin, A.; Validov, S.; Sprenger, G.A. Escherichia coli Reporter Strains Allow for the In Vivo Evaluation of Recombinant Elongation Factor Protein (EF-P). Appl. Microbiol. 2024, 4, 1335-1347. https://doi.org/10.3390/applmicrobiol4030092
Trachtmann N, Bikmullin A, Validov S, Sprenger GA. Escherichia coli Reporter Strains Allow for the In Vivo Evaluation of Recombinant Elongation Factor Protein (EF-P). Applied Microbiology. 2024; 4(3):1335-1347. https://doi.org/10.3390/applmicrobiol4030092
Chicago/Turabian StyleTrachtmann, Natalia, Aydar Bikmullin, Shamil Validov, and Georg A. Sprenger. 2024. "Escherichia coli Reporter Strains Allow for the In Vivo Evaluation of Recombinant Elongation Factor Protein (EF-P)" Applied Microbiology 4, no. 3: 1335-1347. https://doi.org/10.3390/applmicrobiol4030092
APA StyleTrachtmann, N., Bikmullin, A., Validov, S., & Sprenger, G. A. (2024). Escherichia coli Reporter Strains Allow for the In Vivo Evaluation of Recombinant Elongation Factor Protein (EF-P). Applied Microbiology, 4(3), 1335-1347. https://doi.org/10.3390/applmicrobiol4030092







