New-Onset Diabetes Mellitus after COVID-19: Combined Effects of SARS-CoV-2 Variants, Molecular Mimicry, and m6A RNA Methylation
Abstract
:1. Introduction
2. Molecular Mechanisms Involved in NODAC Development
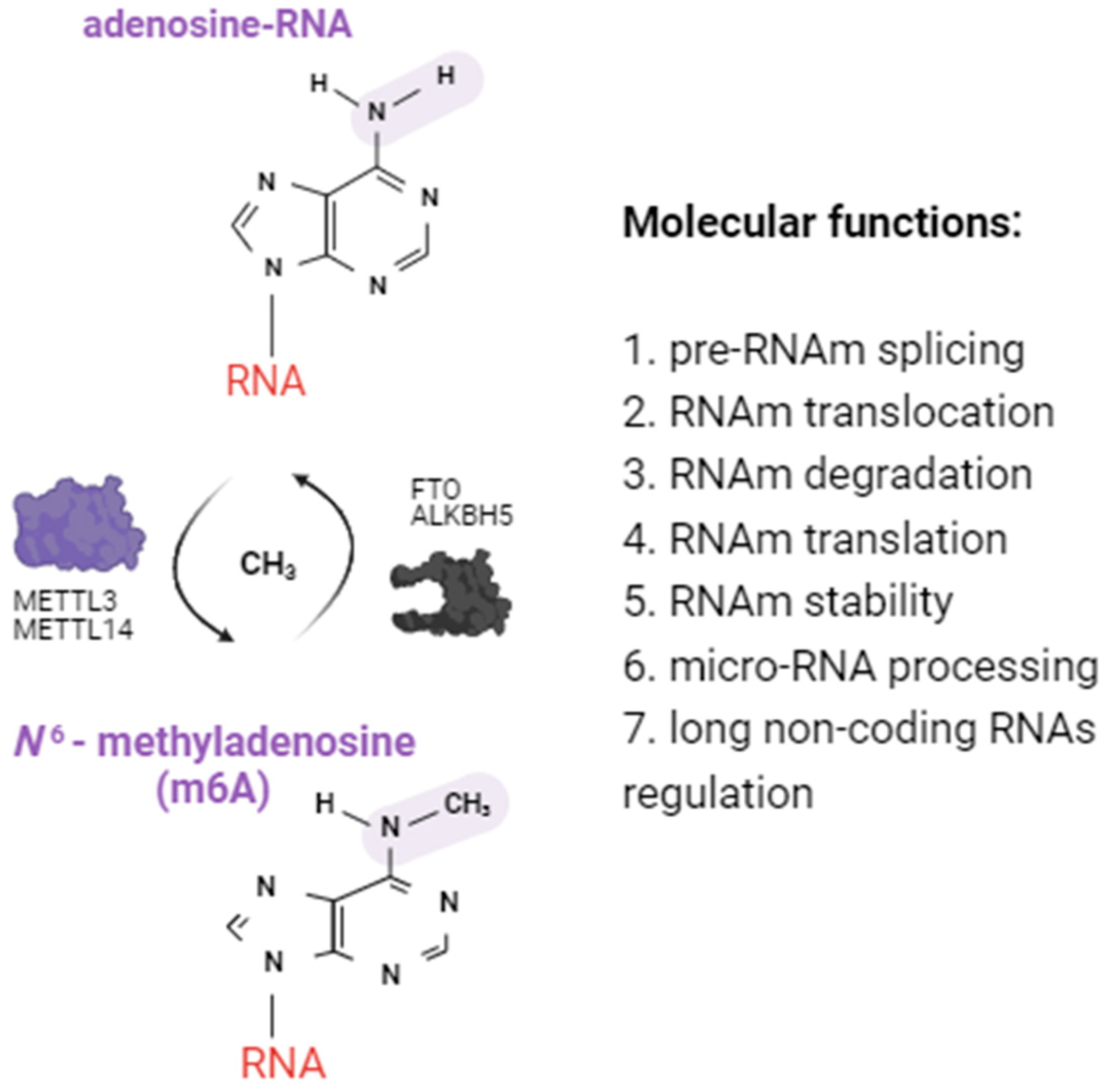
2.1. Alterations of m6A RNA Methylation Patterns
2.1.1. Alterations in Immune Cells from Diabetic Patients
2.1.2. Alterations in Beta Cells from Diabetic Patients
2.1.3. Alterations in m6A Methylation Induced by mRNA Vaccines
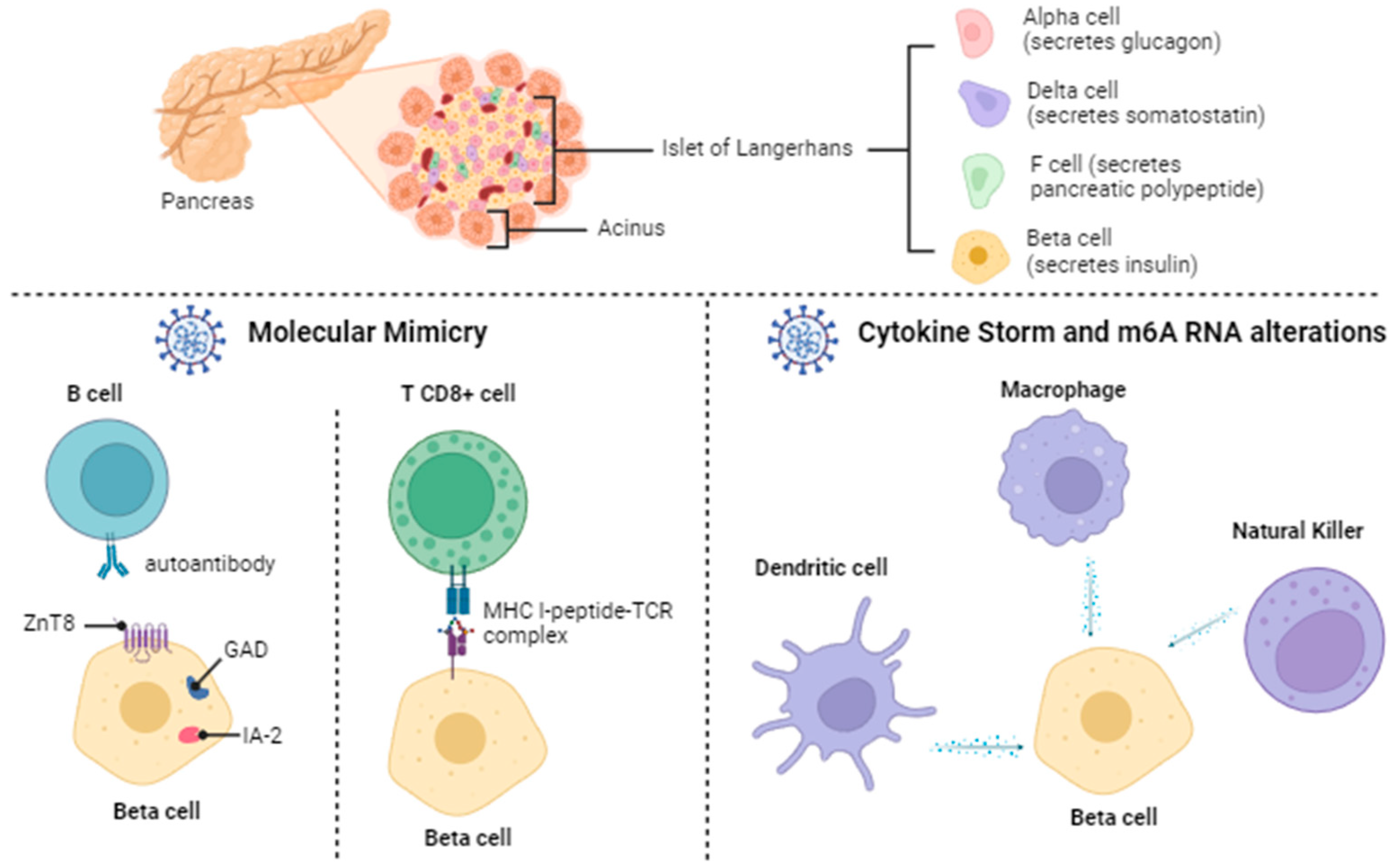
2.2. Diabetes Mellitus: Beta Pancreatic Cell Destruction and Dysfunction
2.3. Direct SARS-CoV-2 Invasion of Beta Cells
2.4. Immune Responses (Autoimmunity and Inflammation) Induced by SARS-CoV-2
2.4.1. Molecular Mimicry
2.4.2. Cytokine Storm
2.5. Alterations in m6A Methylation by Different SARS-CoV-2 Variants
3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef]
- Becerra-Flores, M.; Cardozo, T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020, 74, e13525. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, Y.; Tang, X.; He, D. The Disease Severity and Clinical Outcomes of the SARS-CoV-2 Variants of Concern. Front. Public Health. 2021, 9, 775224. [Google Scholar] [CrossRef]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Haider, N.; Abbasi, A.F.; Jaferi, U.; Prakash, S.; Balendra, V. The emerging SARS-CoV-2 variants of concern. Ther. Adv. Infect. Dis. 2021, 8, 20499361211024372. [Google Scholar] [CrossRef]
- CDC—Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html#anchor_1632237683347 (accessed on 3 January 2024).
- Álvarez-Santacruz, C.; Tyrkalska, S.D.; Candel, S. The Microbiota in Long COVID. Int. J. Mol. Sci. 2024, 25, 1330. [Google Scholar] [CrossRef] [PubMed]
- Baj, J.; Karakuła-Juchnowicz, H.; Teresiński, G.; Buszewicz, G.; Ciesielka, M.; Sitarz, R.; Forma, A.; Karakuła, K.; Flieger, W.; Portincasa, P.; et al. COVID-19: Specific and Non-Specific Clinical Manifestations and Symptoms: The Current State of Knowledge. J. Clin. Med. 2020, 9, 1753. [Google Scholar] [CrossRef]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-term cardiovascular outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef]
- Kedor, C.; Freitag, H.; Meyer-Arndt, L.; Wittke, K.; Hanitsch, L.G.; Zoller, T.; Steinbeis, F.; Haffke, M.; Rudolf, G.; Heidecker, B.; et al. A prospective observational study of post-COVID-19 chronic fatigue syndrome following the first pandemic wave in Germany and biomarkers associated with symptom severity. Nat. Commun. 2022, 13, 101019. [Google Scholar] [CrossRef]
- Antar, S.A.; Ashour, N.A.; Sharaky, M.; Khattab, M.; Ashour, N.A.; Zaid, R.T.; Roh, E.J.; Elkamhawy, A.; Al-Karmalawy, A.A. Diabetes mellitus: Classification, mediators, and complications; A gate to identify potential targets for the development of new effective treatments. Biomed. Pharmacother. 2023, 168, 115734. [Google Scholar] [CrossRef] [PubMed]
- Boddu, S.K.; Aurangabadkar, G.; Kuchay, M.S. New onset diabetes, type 1 diabetes and COVID-19. Diabetes Metab. Syndr. 2020, 14, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Pantea-Stoian, A.; Bica, I.C.; Salmen, T.; Al Mahmeed, W.; Al-Rasadi, K.; Al-Alawi, K.; Banach, M.; Banerjee, Y.; Ceriello, A.; Cesur, M.; et al. New-Onset Diabetes Mellitus in COVID-19: A Scoping Review. Diabetes Ther. 2024, 15, 33–60. [Google Scholar] [CrossRef] [PubMed]
- Ayoubkhani, D.; Khunti, K.; Nafilyan, V.; Maddox, T.; Humberstone, B.; Diamond, I.; Banerjee, A. Post-covid syndrome in individuals admitted to hospital with COVID-19: Retrospective cohort study. BMJ 2021, 372, n693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, S.; Dong, C.; Guo, S.; Jia, W.; Jiang, Y.; Wang, C.; Zhou, M.; Gong, Y. A bibliometric analysis of RNA methylation in diabetes mellitus and its complications from 2002 to 2022. Front. Endocrinol. 2022, 13, 997034. [Google Scholar] [CrossRef] [PubMed]
- Chourasia, P.; Goyal, L.; Kansal, D.; Roy, S.; Singh, R.; Mahata, I.; Sheikh, A.B.; Shekhar, R. Risk of New-Onset Diabetes Mellitus as a Post-COVID-19 Condition and Possible Mechanisms: A Scoping Review. J. Clin. Med. 2023, 12, 1159. [Google Scholar] [CrossRef]
- Raveendran, A.V. COVID-19 and diabetes mellitus: Proposed diagnostic criteria for new-onset diabetes after COVID. Int. J. Diabetes Technol. 2022, 3, 101–103. [Google Scholar] [CrossRef]
- Ghosh, A.; Anjana, R.M.; Shanthi-Rani, C.S.; Jeba-Rani, S.; Gupta, R.; Jha, A.; Gupta, V.; Kuchay, M.S.; Luthra, A.; Durrani, S.; et al. Glycemic parameters in patients with new-onset diabetes during COVID-19 pandemic are more severe than in patients with new-onset diabetes before the pandemic: NOD COVID India Study. Diabetes Metab. Syndr. 2021, 15, 215–220. [Google Scholar] [CrossRef]
- Rathmann, W.; Kuss, O.; Kostev, K. Incidence of newly diagnosed diabetes after COVID-19. Diabetologia 2022, 65, 949–954. [Google Scholar] [CrossRef]
- Qeadan, F.; Tingey, B.; Egbert, J.; Pezzolesi, M.G.; Burge, M.R.; Peterson, K.A.; Honda, T. The associations between COVID-19 diagnosis, type 1 diabetes, and the risk of diabetic ketoacidosis: A nationwide cohort from the US using the Cerner Real-World Data. PLoS ONE 2022, 17, e0266809. [Google Scholar] [CrossRef]
- Xie, Y.; Al-Aly, Z. Risks and burdens of incident diabetes in long COVID: A cohort study. Lancet Diabetes Endocrinol. 2022, 10, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Ssentongo, P.; Zhang, Y.; Witmer, L.; Chinchilli, V.M.; Ba, D.M. Association of COVID-19 with diabetes: A systematic review and meta-analysis. Sci. Rep. 2022, 12, 1–8. [Google Scholar] [CrossRef]
- Lai, H.; Yang, M.; Sun, M.; Pan, B.; Wang, Q.; Wang, J.; Tian, J.; Ding, G.; Yang, K.; Song, X.; et al. Risk of incident diabetes after COVID-19 infection: A systematic review and meta-analysis. Metabolism 2022, 137, 155330. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Wang, Z.; Liu, N.; He, L.; Zhang, H. Increased risk of new-onset diabetes in patients with COVID-19: A systematic review and meta-analysis. Front. Public Health 2023, 11, 1170156. [Google Scholar] [CrossRef] [PubMed]
- Zanardo, V.; Tortora, D.; Sandri, A.; Severino, L.; Mesirca, P.; Straface, G. COVID-19 pandemic: Impact on gestational diabetes mellitus prevalence. Diabetes Res. Clin. Pract. 2022, 183, 109149. [Google Scholar] [CrossRef] [PubMed]
- Rhou, Y.J.J.; Elhindi, J.; Melov, S.J.; Cheung, N.W.; Pasupathy, D. Western Sydney COVID-19 Pregnancy Study Group. Indirect effects of the COVID-19 pandemic on risk of gestational diabetes and factors contributing to increased risk in a multiethnic population: A retrospective cohort study. BMC Pregnancy Childbirth 2023, 23, 341. [Google Scholar] [CrossRef] [PubMed]
- Maharat, M.; Sajjadi, S.F.; Moosavian, S.P. Changes in dietary habits and weight status during the COVID-19 pandemic and its association with socioeconomic status among Iranians adults. Front. Public Health 2023, 10, 1080589. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Khandpur, N.; Desjardins, C.; Wang, L.; Monteiro, C.A.; Rossato, S.L.; Fung, T.T.; Manson, J.E.; Willett, W.C.; Rimm, E.B.; et al. Ultra-Processed Food Consumption and Risk of Type 2 Diabetes: Three Large Prospective U.S. Cohort Studies. Diabetes Care 1 2023, 46, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, S.; Arora, B.; Dutta, K.; Ghosh, A.; Sinha, B.; Misra, A. Increase in the risk of type 2 diabetes during lockdown for the COVID19 pandemic in India: A cohort analysis. Diabetes Metab. Syndr. 2020, 14, 949–952. [Google Scholar] [CrossRef]
- Silva, F.M.; Duarte-Mendes, P.; Carvalho, E.; Soares, C.M.; Farinha, C.; Serrano, J.; Paulo, R.; Massart, A.; Rodrigues, R.N.; Teixeira, A.M.; et al. Effects of combined training during the COVID-19 pandemic on metabolic health and quality of life in sedentary workers: A randomized controlled study. Front. Public Health 2022, 10, 1040714. [Google Scholar] [CrossRef]
- Kim, S.H.; Arora, I.; Hsia, D.S.; Knowler, W.C.; LeBlanc, E.; Mylonakis, E.; Pratley, R.; Pittas, A.G. New-Onset Diabetes After COVID-19. J. Clin. Endocrinol. Metab. 2023, 108, e1164–e1174. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.C.; Cao, H.D.; Lian, X.F.; Wu, P.X.; Zhang, F.; Zhang, H.; Lu, D.H. Molecular mechanisms of noncoding RNA and epigenetic regulation in obesity with consequent diabetes mellitus development. World J. Diabetes 2023, 14, 1621–1631. [Google Scholar] [CrossRef] [PubMed]
- Pan, T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem. Sci. 2013, 38, 204–209. [Google Scholar] [CrossRef] [PubMed]
- De Jesus, D.F.; Zhang, Z.; Kahraman, S.; Brown, N.K.; Chen, M.; Hu, J.; Gupta, M.K.; He, C.; Kulkarni, R.N. m6A mRNA Methylation Regulates Human β-Cell Biology in Physiological States and in Type 2 Diabetes. Nat. Metab. 2019, 1, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Batista-Roche, J.L.; Gómez-Gil, B.; Lund, G.; Berlanga-Robles, C.A.; García-Gasca, A. Global m6A RNA Methylation in SARS-CoV-2 Positive Nasopharyngeal Samples in a Mexican Population: A First Approximation Study. Epigenomes 2022, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Zhou, J.; Duan, L.; Wang, H.; Xu, S.; Cao, Y. m6A methylation: A potential key player in understanding and treating COVID-2019 infection. Cell Death Discov. 2023, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Luo, S.; Sun, X.; Li, J.; Pang, H.; Zhou, J.; Zhou, Y.; Shi, X.; Li, X.; et al. The m6A methylation profiles of immune cells in type 1 diabetes mellitus. Front. Immunol. 2022, 13, 1030728. [Google Scholar] [CrossRef]
- Oldstone, M.B.A.; Schwimmbeck, P.; Dyrberg, T.; Fujinami, R. Mimicry by Virus of Host Molecules: Implications for Autoimmune Disease. In Progress in Immunology VI; Cinader, B., Miller, R.G., Eds.; Academic Press Inc.: Orlando, FL, USA, 1986; pp. 787–795. [Google Scholar] [CrossRef]
- Cusick, M.F.; Libbey, J.E.; Fujinami, R.S. Molecular mimicry as a mechanism of autoimmune disease. Clin. Rev. Allergy Immunol. 2012, 42, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Coppieters, K.T.; Wiberg, A.; von Herrath, M.G. Viral infections and molecular mimicry in type 1 diabetes. APMIS 2012, 120, 941–949. [Google Scholar] [CrossRef]
- Kovačić, D.; Jotanović, J.; Laković, J. The possible role of molecular mimicry in SARS-CoV-2-mediated autoimmunity: An immunobiochemical basis. J. Med. Sci. 2021, 90, 137–156. [Google Scholar] [CrossRef]
- Kanduc, D.; Shoenfeld, Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: Implications for the vaccine. Immunol. Res. 2020, 68, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Gammazza, A.M.; Dieli, F.; de Macario, E.C.; Macario, A.J. Does SARS-CoV-2 Trigger Stress-InducedAutoimmunity by Molecular Mimicry? A Hypothesis. J. Clin. Med. 2020, 9, 2038. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Shen, F.; Huang, W.; Qin, S.; Huang, J.T.; Sergi, C.; Yuan, B.F.; Liu, S.M. Glucose is involved in the dynamic regulation of M6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab. 2019, 104, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.; Sun, X.; Wu, Y.; Chen, Z. METTL3 is required for maintaining β-cell function. Metabolism 2021, 116, 154702. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Luo, G.; Sun, J.; Men, L.; Ye, H.; He, C.; Ren, D. METTL14 is essential for β-cell survival and insulin secretion. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2138–2148. [Google Scholar] [CrossRef]
- Cheng, Y.; Yao, X.M.; Zhou, S.M.; Sun, Y.; Meng, X.J.; Wang, Y.; Xing, Y.J.; Wan, S.J.; Hua, Q. The m6A Methyltransferase METTL3 Ameliorates Methylglyoxal-Induced Impairment of Insulin Secretion in Pancreatic β Cells by Regulating MafA Expression. Front. Endocrinol. 2022, 13, 1664–2392. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.A.; Morgan, N.G. Conditional expression of the FTO gene product in rat INS-1 cells reveals its rapid turnover and a role in the profile of glucose-induced insulin secretion. Clin. Sci. 2011, 120, 403–413. [Google Scholar] [CrossRef]
- Fan, H.Q.; He, W.; Xu, K.F.; Wang, Z.X.; Xu, X.Y.; Chen, H. FTO Inhibits Insulin Secretion and Promotes NF-κB Activation through Positively Regulating ROS Production in Pancreatic β cells. PLoS ONE 2015, 10, e0127705. [Google Scholar] [CrossRef]
- He, Y.F.; Ouyang, J.; Hu, X.D.; Wu, N.; Jiang, Z.G.; Bian, N.; Wang, J. Correlation between COVID-19 vaccination and diabetes mellitus: A systematic review. World J. Diabetes. 2023, 14, 892–918. [Google Scholar] [CrossRef]
- Alsudais, A.S.; Alkanani, R.S.; Fathi, A.B.; Almuntashiri, S.S.; Jamjoom, J.N.; Alzhrani, M.A.; Althubaiti, A.; Radi, S. Autoimmune diabetes mellitus after COVID-19 vaccination in adult population: A systematic review of case reports. BMC Endocr. Disord. 2023, 23, 164. [Google Scholar] [CrossRef]
- Moon, H.; Suh, S.; Park, M.K. Adult-Onset Type 1 Diabetes Development Following COVID-19 mRNA Vaccination. J. Korean Med. Sci. 2023, 38, e12. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Lui, D.T.W.; Chung, M.S.H.; Au, I.C.H.; Lai, F.T.T.; Wan, E.Y.F.; Chui, C.S.L.; Li, X.; Cheng, F.W.T.; Cheung, C.L.; et al. Incidence of diabetes following COVID-19 vaccination and SARS-CoV-2 infection in Hong Kong: A population-based cohort study. PLoS Med. 2023, 20, e1004274. [Google Scholar] [CrossRef] [PubMed]
- Vera, F.; Jofré, P.; Rodríguez, D.; Lagos, R.; Quiroga, T.; Grassi, B.; Pollak, F. Marcadores inmunológicos de diabetes tipo 1: Análisis de 4.164 perfiles en un centro de salud de Chile. Rev. Méd. Chile 2021, 149, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Hayes-Dorado, J.P. Diabetes mellitus tipo 1. Rev. Bol. Ped. 2008, 47, 90–96. Available online: http://www.scielo.org.bo/scielo.php?script=sci_arttext&pid=S1024-06752008000200006 (accessed on 15 February 2024).
- Kokuina, E.; Rozada Galardy, L.; Vilar MD, L.N.; Tamargo Barbeito, T.O.; Infante Amoros, A.; Dalas Giber, M. Autoanticuerpos anti-insulina en la diabetes tipo 1. Rev. Cuba. Med. 2022, 61, e2637. Available online: https://pesquisa.bvsalud.org/portal/resource/e/biblio-1408993 (accessed on 15 February 2024).
- Cerf, M.E. Beta cell dysfunction and insulin resistance. Front. Endocrinol. 2013, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Reiterer, M.; Rajan, M.; Gómez-Banoy, N.; Lau, J.D.; Gomez-Escobar, L.G.; Ma, L.; Gilani, A.; Alvarez-Mulett, S.; Sholle, E.T.; Chandar, V.; et al. Hyperglycemia in acute COVID-19 is characterized by insulin resistance and adipose tissue infectivity by SARS-CoV-2. Cell Metab. 2021, 33, 2174–2188.e5. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Liu, C.; Peng, J.; Li, Z.; Li, F.; Wang, J.; Hu, A.; Peng, M.; Huang, K.; Fan, D.; et al. COVID-19 induces new-onset insulin resistance and lipid metabolic dysregulation via regulation of secreted metabolic factors. Signal Transduct. Target. Ther. 2021, 6, 1–12. [Google Scholar] [CrossRef]
- Kleinwechter, H.J.; Weber, K.S.; Liedtke, T.P.; Schäfer-Graf, U.; Groten, T.; Rüdiger, M.; Pecks, U. COVID-19, Pregnancy, and Diabetes Mellitus. Z. Geburtshilfe Neonatol. 2024, 228, 17–31. [Google Scholar] [CrossRef]
- Modzelewski, R.; Stefanowicz-Rutkowska, M.M.; Matuszewski, W.; Bandurska-Stankiewicz, E.M. Gestational Diabetes Mellitus-Recent Literature Review. J. Clin. Med. 2022, 11, 5736. [Google Scholar] [CrossRef]
- Taneera, J.; El-Huneidi, W.; Hamad, M.; Mohammed, A.K.; Elaraby, E.; Hachim, M.Y. Expression Profile of SARS-CoV-2 Host Receptors in Human Pancreatic Islets Revealed Upregulation of ACE2 in Diabetic Donors. Biology 2020, 9, 215. [Google Scholar] [CrossRef] [PubMed]
- Fignani, D.; Licata, G.; Brusco, N.; Nigi, L.; Grieco, G.E.; Marselli, L.; Overbergh, L.; Gysemans, C.; Colli, M.L.; Marchetti, P.; et al. SARS-CoV-2 Receptor Angiotensin I-Converting Enzyme Type 2 (ACE2) Is Expressed in Human Pancreatic β-Cells and in the Human Pancreas Microvasculature. Front. Endocrinol. 2020, 11, 596898. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Long, X.; Zhang, B.; Zhang, W.; Chen, X.; Zhang, Z. ACE2 expression in pancreas may cause pancreatic damage after SARS-CoV-2 infection. Clin. Gastroenterol. Hepatol. 2020, 18, 2128–2130. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Lee, I.T.; Cheng, R.; Nakayama, T.; Jiang, S.; Demeter, J.; Bevacqua, R.J.; Chang, C.A.; et al. SARS-CoV-2 infects human pancreatic β cells and elicits β cell impairment. Cell Metab. 2021, 33, 1565–1576.e5. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.A.; Groß, R.; Conzelmann, C.; Krüger, J.; Merle, U.; Steinhart, J.; Weil, T.; Koepke, L.; Bozzo, C.P.; Read, C.; et al. SARS-CoV-2 infects and replicates in cells of the human endocrine and exocrine pancreas. Nat. Metab. 2021, 3, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Rubino, F.; Amiel, S.A.; Zimmet, P.; Alberti, G.; Bornstein, S.; Eckel, R.H. New-onset diabetes in COVID-19. N. Engl. J. Med. 2020, 383, 789–790. [Google Scholar] [CrossRef]
- Lima-Martínez, M.M.; Carrera Boada, C.; Madera-Silva, M.D.; Marín, W.; Contreras, M. COVID-19 and diabetes: A bidirectional relationship. COVID-19 y diabetes mellitus: Una relación bidireccional. Clin. Investig. Arterioscler. 2021, 33, 151–157. [Google Scholar] [CrossRef]
- Mine, K.; Nagafuchi, S.; Mori, H.; Takahashi, H.; Anzai, K. SARS-CoV-2 Infection and Pancreatic β Cell Failure. Biology 2022, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Morais da Silva, M.; Lira de Lucena, A.S.; Paiva Júnior, S.S.L.; Florêncio De Carvalho, V.M.; Santana de Oliveira, P.S.; da Rosa, M.M.; Barreto de Melo Rego, M.J.; Pitta, M.G.D.R.; Pereira, M.C. Cell death mechanisms involved in cell injury caused by SARS-CoV-2. Rev. Med. Virol. 2022, 32, e2292. [Google Scholar] [CrossRef]
- Ben Nasr, M.; D‘Addio, F.; Montefusco, L.; Usuelli, V.; Loretelli, C.; Rossi, A.; Pastore, I.; Abdelsalam, A.; Maestroni, A.; Dell’Acqua, M.; et al. Indirect and Direct Effects of SARS-CoV-2 on Human Pancreatic Islets. Diabetes 2022, 71, 1579–1590. [Google Scholar] [CrossRef]
- Saberzadeh-Ardestani, B.; Karamzadeh, R.; Basiri, M.; Hajizadeh-Saffar, E.; Farhadi, A.; Shapiro, A.M.J.; Tahamtani, Y.; Baharvand, H. Type 1 Diabetes Mellitus: Cellular and Molecular Pathophysiology at A Glance. Cell J. 2018, 20, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Gavkare, A.M.; Nanaware, N.; Rayate, A.S.; Mumbre, S.; Nagoba, B.S. COVID-19 associated diabetes mellitus: A review. World J. Diabetes 2022, 13, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Moody, R.; Wilson, K.L.; Boer, J.C.; Holien, J.K.; Flanagan, K.L.; Jaworowski, A.; Plebanski, M. Predicted B Cell Epitopes Highlight the Potential for COVID-19 to Drive Self- Reactive Immunity. Front. Bioinform. 2021, 1, 709533. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Herrán, M.; Ramírez-Santana, C.; Leung, P.S.C.; Anaya, J.M.; Ridgway, W.M.; Gershwin, M.E. Molecular mimicry and autoimmunity in the time of COVID-19. J. Autoimmun. 2023, 139, 103070. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Castilla, J.; Stebliankin, V.; Baral, P.; Balbin, C.A.; Sobhan, M.; Cickovski, T.; Mondal, A.M.; Narasimhan, G.; Chapagain, P.; Mathee, K.; et al. Potential Autoimmunity Resulting from Molecular Mimicry between SARS-CoV-2 Spike and Human Proteins. Viruses 2022, 14, 1415. [Google Scholar] [CrossRef]
- Laxminarayana, D. Molecular insights into onset of autoimmunity in SARS-CoV-2 infected patients. Rheumatol. Autoimmun. 2022, 2, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Churilov, L.P.; Kanduc, D.; Ryabkova, V.A. COVID-19: Adrenal response and molecular mimicry. Isr. Med. Assoc. J. 2021, 23, 618–619. Available online: https://www.ima.org.il/MedicineIMAJ/viewarticle.aspx?year=2021&month=10&page=618 (accessed on 15 February 2024).
- Churilov, L.P.; Normatov, M.G.; Utekhin, V.J. Molecular Mimicry between SARS-CoV-2 and Human Endocrinocytes: A Prerequisite of Post-COVID-19 Endocrine Autoimmunity? Pathophysiology 2022, 29, 486–494. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, G.L.V.; Oliveira, C.N.S.; Pinzan, C.F.; de Salis, L.V.V.; Cardoso, C.R.B. Microbiota Modulation of the Gut-Lung Axis in COVID-19. Front. Immunol. 2021, 12, 635471. [Google Scholar] [CrossRef]
- Papachristou, S.; Stamatiou, I.; Stoian, A.P.; Papanas, N. New-Onset Diabetes in COVID-19: Time to Frame Its Fearful Symmetry. Diabetes Ther. 2021, 12, 461–464. [Google Scholar] [CrossRef]
- Galleri, L.; Sebastiani, G.; Vendrame, F.; Grieco, F.A.; Spagnuolo, I.; Dotta, F. Viral Infections and Diabetes. Adv. Exp. Med. Biol. 2013, 771, 252–271. [Google Scholar] [CrossRef]
- Srivastava, A.; Rockman-Greenberg, C.; Sareen, N.; Lionetti, V.; Dhingra, S. An insight into the mechanisms of COVID-19, SARS-CoV-2 infection severity concerning β-cell survival and cardiovascular conditions in diabetic patients. Mol. Cell Biochem. 2022, 477, 1681–1695. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Chu, C.Q. Th17 Cells in Type 1 Diabetes: Role in the Pathogenesis and Regulation by Gut Microbiome. Mediators Inflamm. 2015, 2015, 638470. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.; Bakery, H.H.; Allam, G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed. Pharmacother. 2018, 101, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Chakraborty, R.; Kalita, P.; Pathak, M.P. Diabetes mellitus and COVID-19: Understanding the association in light of current evidence. World J. Clin. Cases 2021, 9, 8327–8339. [Google Scholar] [CrossRef]
- Mahrooz, A.; Muscogiuri, G.; Buzzetti, R.; Maddaloni, E. The complex combination of COVID-19 and diabetes: Pleiotropic changes in glucose metabolism. Endocrine 2021, 72, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Rangu, R.; Wander, P.L.; Zraika, S. Does diabetes risk after SARS-CoV-2 infection depend on the viral variant? Diabetes Res. Clin. Pract. 2022, 191, 110054. [Google Scholar] [CrossRef]
- Vaid, R.; Mendez, A.; Thombare, K.; Burgos-Panadero, R.; Robinot, R.; Fonseca, B.F.; Gandasi, N.R.; Ringlander, J.; Hassan Baig, M.; Dong, J.J.; et al. Global loss of cellular m6A RNA methylation following infection with different SARS-CoV-2 variants. Genome Res. 2023, 33, 299–313. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batista-Roche, J.L.; Mirabent-Casals, M.; Manzanares, D.; Lund, G.; García-Gasca, A. New-Onset Diabetes Mellitus after COVID-19: Combined Effects of SARS-CoV-2 Variants, Molecular Mimicry, and m6A RNA Methylation. COVID 2024, 4, 481-494. https://doi.org/10.3390/covid4040032
Batista-Roche JL, Mirabent-Casals M, Manzanares D, Lund G, García-Gasca A. New-Onset Diabetes Mellitus after COVID-19: Combined Effects of SARS-CoV-2 Variants, Molecular Mimicry, and m6A RNA Methylation. COVID. 2024; 4(4):481-494. https://doi.org/10.3390/covid4040032
Chicago/Turabian StyleBatista-Roche, Jorge Luis, Marian Mirabent-Casals, Dahis Manzanares, Gertrud Lund, and Alejandra García-Gasca. 2024. "New-Onset Diabetes Mellitus after COVID-19: Combined Effects of SARS-CoV-2 Variants, Molecular Mimicry, and m6A RNA Methylation" COVID 4, no. 4: 481-494. https://doi.org/10.3390/covid4040032





