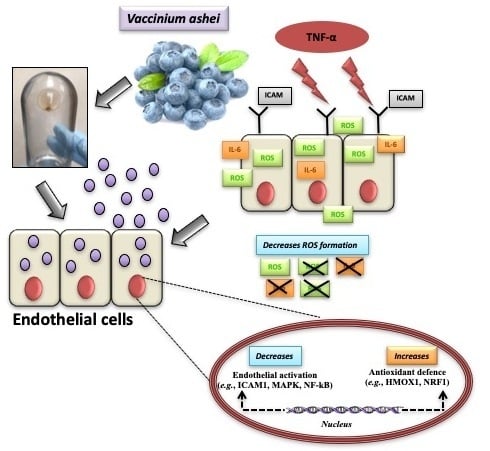
Blueberry-Derived Exosome-Like Nanoparticles Counter the Response to TNF-α-Induced Change on Gene Expression in EA.hy926 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Characterization of B-ELNs
2.2. EA.hy926 Cell Culture and Experimental Design
2.3. B-ELNs Cellular Transport and Time Course Study
2.4. Cell Viability
2.5. Detection of Intracellular Superoxide Anion/Superoxide-Derived ROS with Dihydroethidium (DHE)
2.6. RNA Isolation, qRT-PCR, and Gene Expression Profile
2.7. Functional Enrichment Analysis
2.8. Prediction and Functional Annotation of miRNA Target Genes
2.9. Statistics
3. Results
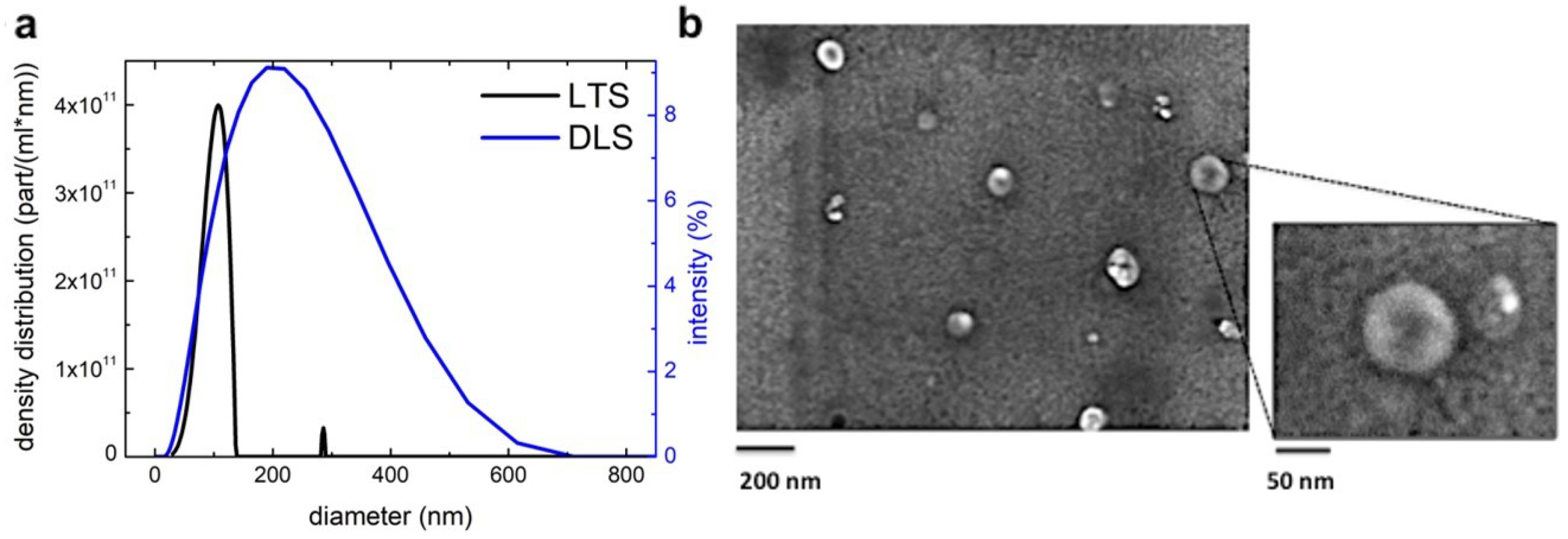
3.1. Blueberry (Vaccinium Ashei) Contains Exosome-Like Nanoparticles
3.2. B-ELNs are Internalized by EA.hy926 in a Dose-Depend Manner
3.3. B-ELNs Protect EA.hy926 from TNF-α-Induced Cytotoxicity and Oxidative Stress
3.4. Effect of B-ELNs on TNF-α Induced mRNA Expression Changes
3.5. miR-156e, miR-162, and miR-319d in B-ELNs Potentially Regulate Mammalian Inflammation-Related Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ELN | exosome-like nanoparticles |
| EVs | extracellular vesicles |
| HMOX1 | heme oxygenase (decycling) 1 |
| IL-6 | interleukin 6 |
| ICAM-1 | Intercellular Adhesion Molecule 1 |
| IL1RL1 | Interleukin 1 receptor-like 1 |
| MAPK | Mitogen-activated protein kinase |
| NAC | N-acetyl-l-cysteine |
| NF-kB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| NRF1 | nuclear respiratory factor 1 |
| PDE7A | phosphodiesterase 7A |
| PTGIS | prostaglandin I2 (prostacyclin) synthase |
| PTGS2 | Prostaglandin-endoperoxide synthase 2/ cyclooxygenase-2 |
| ROS | reactive oxygen species |
| TLR8 | Toll-like receptor 8 |
| TNF-α | tumor necrosis factor alpha |
References
- Yim, N.; Choi, C. Extracellular vesicles as novel carriers for therapeutic molecules. BMB Rep. 2016, 49, 585. [Google Scholar] [CrossRef]
- Maas, S.L.; Breakefield, X.O.; Weaver, A.M. Extracellular vesicles: Unique intercellular delivery vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, 6977. [Google Scholar] [CrossRef] [PubMed]
- Iravani, S.; Varma, R.S. Plant-derived edible nanoparticles and miRNAs: Emerging frontier for therapeutics and targeted drug-delivery. ACS Sustain. Chem. Eng. 2019, 7, 8055–8069. [Google Scholar] [CrossRef]
- Pérez-Bermúdez, P.; Blesa, J.; Soriano, J.M.; Marcilla, A. Extracellular vesicles in food: Experimental evidence of their secretion in grape fruits. Eur. J. Pharm. Sci. 2017, 98, 40–50. [Google Scholar] [CrossRef]
- Liu, B.; Lu, Y.; Chen, X.; Muthuraj, P.G.; Li, X.; Pattabiraman, M.; Zempleni, J.; Kachman, S.D.; Natarajan, S.K.; Yu, J. Protective Role of Shiitake Mushroom-Derived Exosome-Like Nanoparticles in D-Galactosamine and Lipopolysaccharide-Induced Acute Liver Injury in Mice. Nutrients 2020, 12, 477. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 2018, 24, 637–652. [Google Scholar] [CrossRef]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhou, Y.; Yu, J. Exosome-like nanoparticles from ginger rhizomes inhibited NLRP3 inflammasome activation. Mol. Pharm. 2019, 16, 2690–2699. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Zhuang, X.; Wang, Q.; Jiang, H.; Deng, Z.B.; Wang, B.; Zhang, L.; Kakar, S.; Jun, Y.; Miller, D. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol. Nutr. Food Res. 2014, 58, 1561–1573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Viennois, E.; Xu, C.; Merlin, D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers 2016, 4, e1134415. [Google Scholar] [CrossRef]
- Baldini, N.; Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Avnet, S. Exosome-like Nanovesicles Isolated from Citrus limon L. Exert Anti-oxidative Effect. Curr. Pharm. Biotechnol. 2018, 19, 877–885. [Google Scholar] [CrossRef]
- Raimondo, S.; Naselli, F.; Fontana, S.; Monteleone, F.; Dico, A.L.; Saieva, L.; Zito, G.; Flugy, A.; Manno, M.; Di Bella, M.A. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015, 6, 19514. [Google Scholar] [CrossRef]
- Raimondo, S.; Saieva, L.; Cristaldi, M.; Monteleone, F.; Fontana, S.; Alessandro, R. Label-free quantitative proteomic profiling of colon cancer cells identifies acetyl-CoA carboxylase alpha as antitumor target of Citrus limon-derived nanovesicles. J. Proteom. 2018, 173, 1–11. [Google Scholar] [CrossRef]
- Fujita, D.; Arai, T.; Komori, H.; Shirasaki, Y.; Wakayama, T.; Nakanishi, T.; Tamai, I. Apple-derived nanoparticles modulate expression of organic-anion-transporting polypeptide (OATP) 2B1 in Caco-2 cells. Mol. Pharm. 2018, 15, 5772–5780. [Google Scholar] [CrossRef]
- Rome, S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019, 10, 529–538. [Google Scholar] [CrossRef]
- Record, M. Exosome-like nanoparticles from food: Protective nanoshuttles for bioactive cargo. Mol. Ther. 2013, 21, 1294–1296. [Google Scholar] [CrossRef]
- Serraino, I.; Dugo, L.; Dugo, P.; Mondello, L.; Mazzon, E.; Dugo, G.; Caputi, A.P.; Cuzzocrea, S. Protective effects of cyanidin-3-O-glucoside from blackberry extract against peroxynitrite-induced endothelial dysfunction and vascular failure. Life Sci. 2003, 73, 1097–1114. [Google Scholar] [CrossRef]
- Speciale, A.; Anwar, S.; Canali, R.; Chirafisi, J.; Saija, A.; Virgili, F.; Cimino, F. Cyanidin-3-O-glucoside counters the response to TNF-alpha of endothelial cells by activating Nrf2 pathway. Mol. Nutr. Food Res. 2013, 57, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Fratantonio, D.; Speciale, A.; Canali, R.; Natarelli, L.; Ferrari, D.; Saija, A.; Virgili, F.; Cimino, F. Low nanomolar caffeic acid attenuates high glucose-induced endothelial dysfunction in primary human umbilical-vein endothelial cells by affecting NF-κB and Nrf2 pathways. BioFactors 2017, 43, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Fratantonio, D.; Speciale, A.; Molonia, M.; Bashllari, R.; Palumbo, M.; Saija, A.; Cimino, F.; Monastra, G.; Virgili, F. Alpha-lipoic acid, but not di-hydrolipoic acid, activates Nrf2 response in primary human umbilical-vein endothelial cells and protects against TNF-α induced endothelium dysfunction. Arch. Biochem. Biophys. 2018, 655, 18–25. [Google Scholar] [CrossRef]
- Speciale, A.; Cimino, F.; Saija, A.; Canali, R.; Virgili, F. Bioavailability and molecular activities of anthocyanins as modulators of endothelial function. Genes Nutr. 2014, 9, 404. [Google Scholar] [CrossRef]
- Provencher, S.W. A constrained regularization method for inverting data represented by linear algebraic or integral equations. Comput. Phys. Commun. 1982, 27, 213–227. [Google Scholar] [CrossRef]
- De Vos, C.; Deriemaeker, L.; Finsy, R. Quantitative assessment of the conditioning of the inversion of quasi-elastic and static light scattering data for particle size distributions. Langmuir 1996, 12, 2630–2636. [Google Scholar] [CrossRef]
- Li, F.; Schafer, R.; Hwang, C.-T.; Tanner, C.E.; Ruggiero, S.T. High-precision sizing of nanoparticles by laser transmission spectroscopy. Appl. Opt. 2010, 49, 6602–6611. [Google Scholar] [CrossRef]
- De Marcellis, A.; Sarra, A.; Stanchieri, G.D.P.; Bruni, F.; Bordi, F.; Palange, E.; Postorino, P. Balanced Laser Transmission Spectroscopy Based on a Tunable Gain Double Channel LIA for Nanoparticles Detection in Biomedical Applications. In Proceedings of the 2019 IEEE Biomedical Circuits and Systems Conference (BioCAS), Nara, Japan, 17–19 October 2019; pp. 1–4. [Google Scholar]
- Bohren, C.; Huffman, D.; Kam, Z. Book-review-absorption and scattering of light by small particles. Nature 1983, 306, 625. [Google Scholar]
- Anwar, S.; Fratantonio, D.; Ferrari, D.; Saija, A.; Cimino, F.; Speciale, A. Berry anthocyanins reduce proliferation of human colorectal carcinoma cells by inducing caspase-3 activation and p21 upregulation. Mol. Med. Rep. 2016, 14, 1397–1403. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, M.-H. Measurement of reactive oxygen species (ROS) and mitochondrial ROS in AMPK knockout mice blood vessels. In AMPK; Springer: Berlin/Heidelberg, Germany, 2018; pp. 507–517. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Feng, S.; Wang, X.; Long, K.; Luo, Y.; Wang, Y.; Ma, J.; Tang, Q.; Jin, L.; Li, X. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. Peer. J. 2018, 6, e5186. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, 155–159. [Google Scholar] [CrossRef]
- Halasi, M.; Wang, M.; Chavan, T.S.; Gaponenko, V.; Hay, N.; Gartel, A.L. ROS inhibitor N-acetyl-L-cysteine antagonizes the activity of proteasome inhibitors. Biochem. J. 2013, 454, 201–208. [Google Scholar] [CrossRef]
- Miro, X.; Casacuberta, J.M.; Gutierrez-Lopez, M.D.; de Landazuri, M.O.; Puigdomenech, P. Phosphodiesterases 4D and 7A splice variants in the response of HUVEC cells to TNF-alpha(1). Biochem. Biophys. Res. Commun. 2000, 274, 415–421. [Google Scholar] [CrossRef]
- Hoefen, R.J.; Berk, B.C. The role of MAP kinases in endothelial activation. Vasc. Pharmacol. 2002, 38, 271–273. [Google Scholar] [CrossRef]
- Kanda, H.; Kobayashi, K.; Yamanaka, H.; Okubo, M.; Noguchi, K. Microglial TNFalpha Induces COX2 and PGI2 Synthase Expression in Spinal Endothelial Cells during Neuropathic Pain. eNeuro 2017, 4. [Google Scholar] [CrossRef]
- Wohleb, E.S.; Patterson, J.M.; Sharma, V.; Quan, N.; Godbout, J.P.; Sheridan, J.F. Knockdown of interleukin-1 receptor type-1 on endothelial cells attenuated stress-induced neuroinflammation and prevented anxiety-like behavior. J. Neurosci. 2014, 34, 2583–2591. [Google Scholar] [CrossRef]
- Bertok, S.; Wilson, M.R.; Dorr, A.D.; Dokpesi, J.O.; O’Dea, K.P.; Marczin, N.; Takata, M. Characterization of TNF receptor subtype expression and signaling on pulmonary endothelial cells in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 2011, 300, 781–789. [Google Scholar] [CrossRef]
- Sagini, K.; Urbanelli, L.; Buratta, S.; Leonardi, L.; Emiliani, C. Nanovesicles from plants as edible carriers of bioactive compounds. AgroLife Sci. J. 2017, 6, 167–171. [Google Scholar]
- Akuma, P.; Okagu, O.D.; Udenigwe, C.C. Naturally occurring exosome vesicles as potential delivery vehicle for bioactive compounds. Front. Sustain. Food Syst. 2019, 3, 23. [Google Scholar] [CrossRef]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, D.; Speciale, A.; Cristani, M.; Fratantonio, D.; Molonia, M.S.; Ranaldi, G.; Saija, A.; Cimino, F. Cyanidin-3-O-glucoside inhibits NF-kB signalling in intestinal epithelial cells exposed to TNF-α and exerts protective effects via Nrf2 pathway activation. Toxicol. Lett. 2016, 264, 51–58. [Google Scholar] [CrossRef]
- Chen, X.; Andresen, B.T.; Hill, M.; Zhang, J.; Booth, F.; Zhang, C. Role of Reactive Oxygen Species in Tumor Necrosis Factor-alpha Induced Endothelial Dysfunction. Curr. Hypertens. Rev. 2008, 4, 245–255. [Google Scholar] [CrossRef]
- Pan, W.; Yu, H.; Huang, S.; Zhu, P. Resveratrol protects against TNF-α-induced injury in human umbilical endothelial cells through promoting sirtuin-1-induced repression of NF-KB and p38 MAPK. PLoS ONE 2016, 11, e0147034. [Google Scholar] [CrossRef]
- Su, P.; Du, S.; Li, H.; Li, Z.; Xin, W.; Zhang, W. Notoginsenoside R1 inhibits oxidized low-density lipoprotein induced inflammatory cytokines production in human endothelial EA. hy926 cells. Eur. J. Pharmacol. 2016, 770, 9–15. [Google Scholar] [CrossRef]
- Wang, S.; Sarriá, B.; Mateos, R.; Goya, L.; Bravo-Clemente, L. TNF-α-induced oxidative stress and endothelial dysfunction in EA. hy926 cells is prevented by mate and green coffee extracts, 5-caffeoylquinic acid and its microbial metabolite, dihydrocaffeic acid. Int. J. Food Sci. Nutr. 2019, 70, 267–284. [Google Scholar] [CrossRef]
- Zhou, P.; Lu, S.; Luo, Y.; Wang, S.; Yang, K.; Zhai, Y.; Sun, G.; Sun, X. Attenuation of TNF-α-induced inflammatory injury in endothelial cells by ginsenoside Rb1 via inhibiting NF-κB, JNK and p38 signaling pathways. Front. Pharmacol. 2017, 8, 464. [Google Scholar] [CrossRef]
- Fratantonio, D.; Speciale, A.; Ferrari, D.; Cristani, M.; Saija, A.; Cimino, F. Palmitate-induced endothelial dysfunction is attenuated by cyanidin-3-O-glucoside through modulation of Nrf2/Bach1 and NF-kappaB pathways. Toxicol. Lett. 2015, 239, 152–160. [Google Scholar] [CrossRef]
- Lukasik, A.; Zielenkiewicz, P. Plant microRNAs—Novel players in natural medicine? Int. J. Mol. Sci. 2017, 18, 9. [Google Scholar] [CrossRef] [PubMed]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Robertis, M.; Sarra, A.; D’Oria, V.; Mura, F.; Bordi, F.; Postorino, P.; Fratantonio, D. Blueberry-Derived Exosome-Like Nanoparticles Counter the Response to TNF-α-Induced Change on Gene Expression in EA.hy926 Cells. Biomolecules 2020, 10, 742. https://doi.org/10.3390/biom10050742
De Robertis M, Sarra A, D’Oria V, Mura F, Bordi F, Postorino P, Fratantonio D. Blueberry-Derived Exosome-Like Nanoparticles Counter the Response to TNF-α-Induced Change on Gene Expression in EA.hy926 Cells. Biomolecules. 2020; 10(5):742. https://doi.org/10.3390/biom10050742
Chicago/Turabian StyleDe Robertis, Mariangela, Angelo Sarra, Valentina D’Oria, Francesco Mura, Federico Bordi, Paolo Postorino, and Deborah Fratantonio. 2020. "Blueberry-Derived Exosome-Like Nanoparticles Counter the Response to TNF-α-Induced Change on Gene Expression in EA.hy926 Cells" Biomolecules 10, no. 5: 742. https://doi.org/10.3390/biom10050742
APA StyleDe Robertis, M., Sarra, A., D’Oria, V., Mura, F., Bordi, F., Postorino, P., & Fratantonio, D. (2020). Blueberry-Derived Exosome-Like Nanoparticles Counter the Response to TNF-α-Induced Change on Gene Expression in EA.hy926 Cells. Biomolecules, 10(5), 742. https://doi.org/10.3390/biom10050742