The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins
Abstract
:1. Introduction
2. Trp Codon, UGG
3. Trp Biosynthesis: Salient and Unique Features
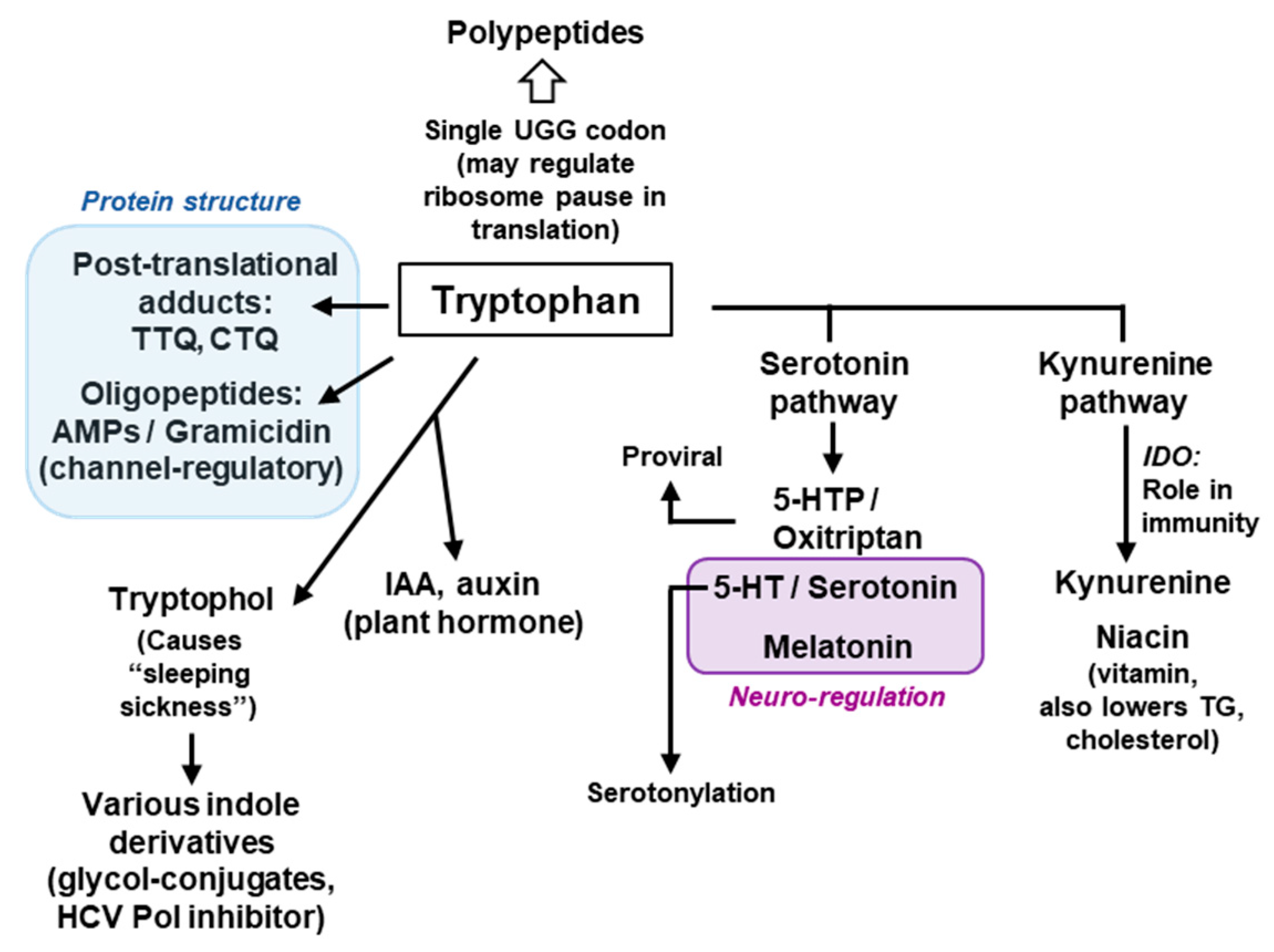
4. Metabolism of Trp
4.1. Trp Degradation Pathways and the Gateway Enzymes
4.2. Secondary Metabolites of Trp
4.2.1. Metabolites of the Serotonin Pathway
4.2.2. Metabolites of the Kynurenine Pathway
4.2.3. Tryptophol and Related Indole Derivatives
4.2.4. Inhibition of Gluconeogenesis by Trp
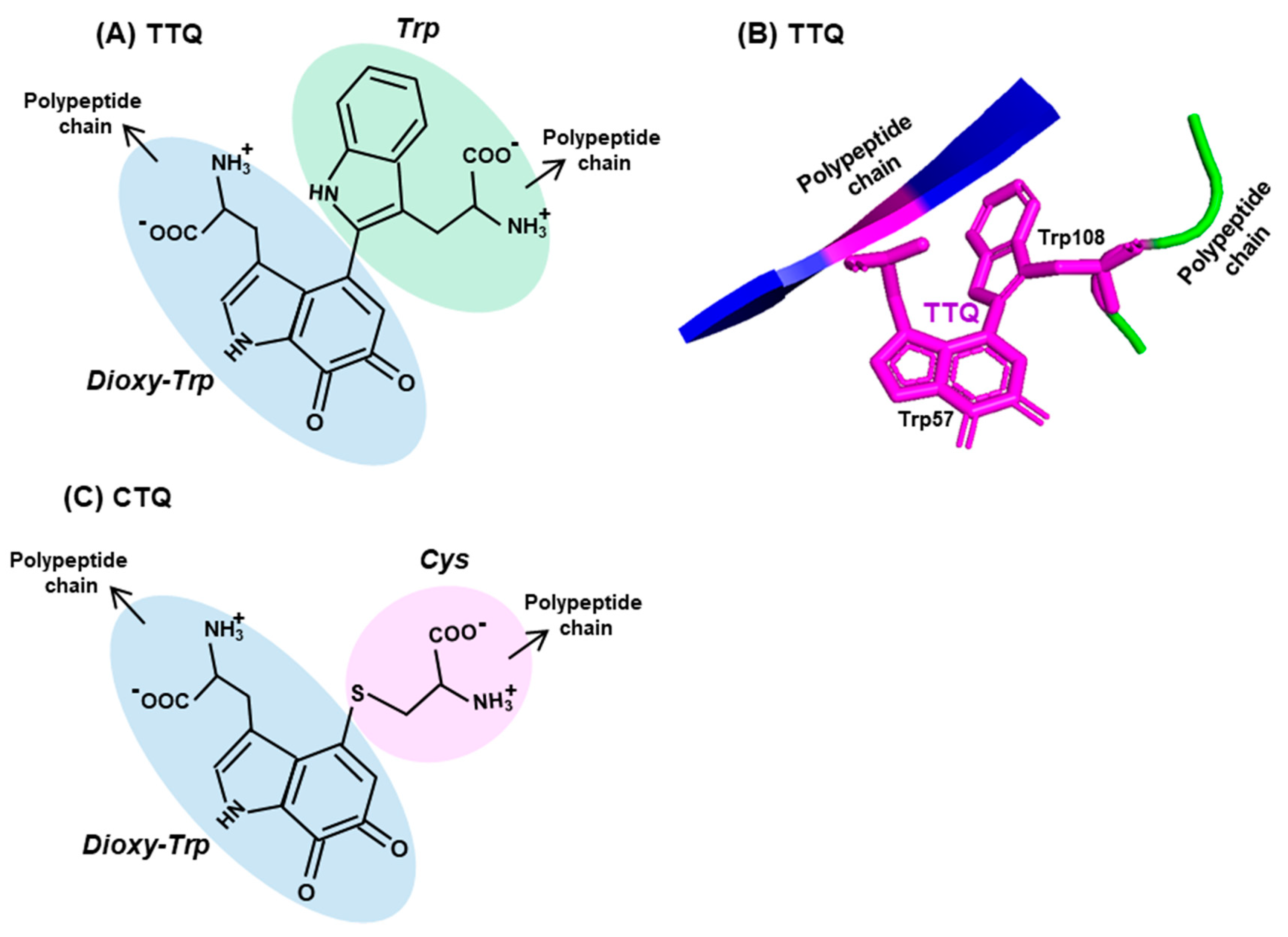
5. Trp Adducts in Proteins
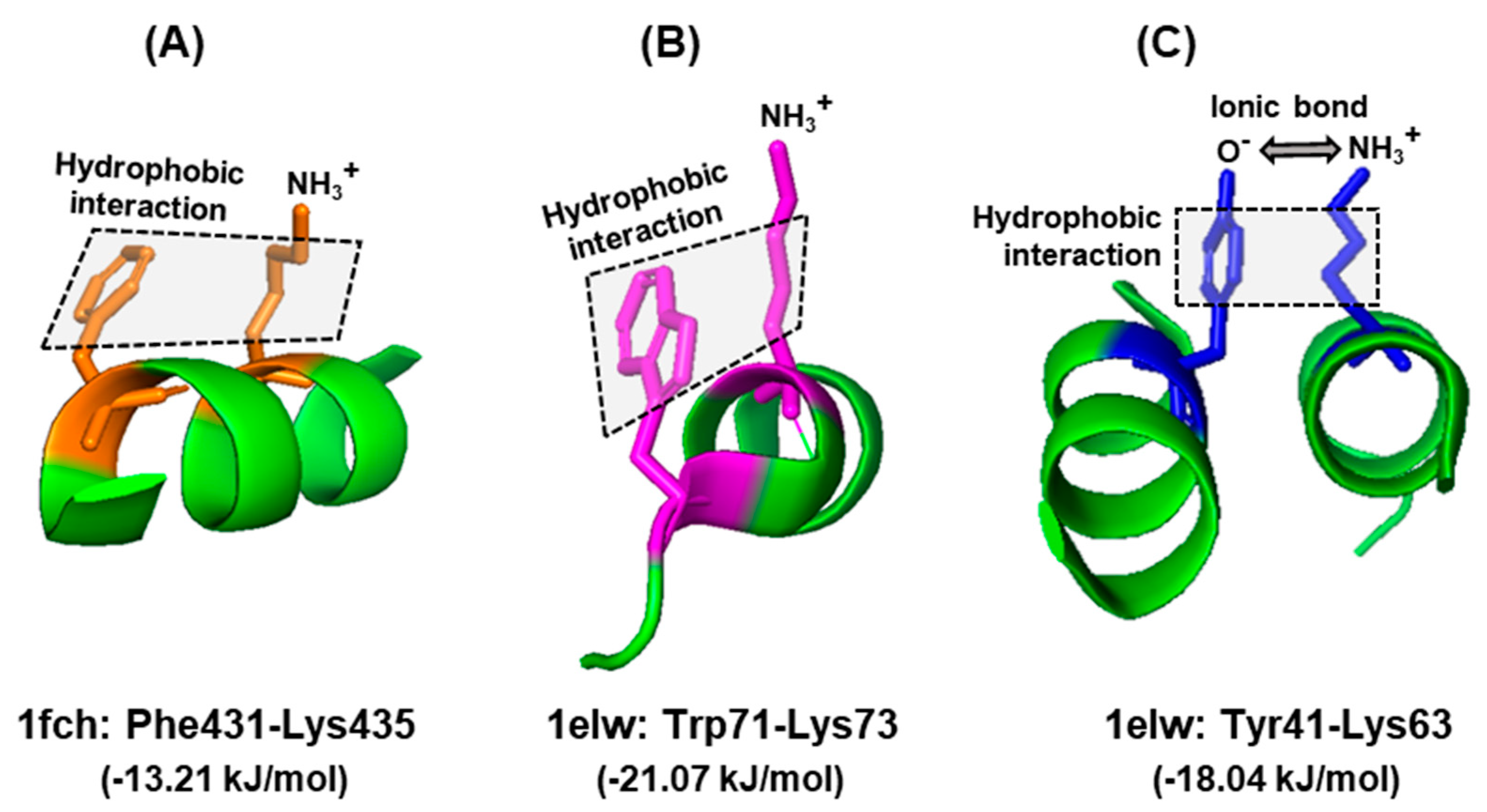
6. Interacting Partners of Trp Residues in A Polypeptide
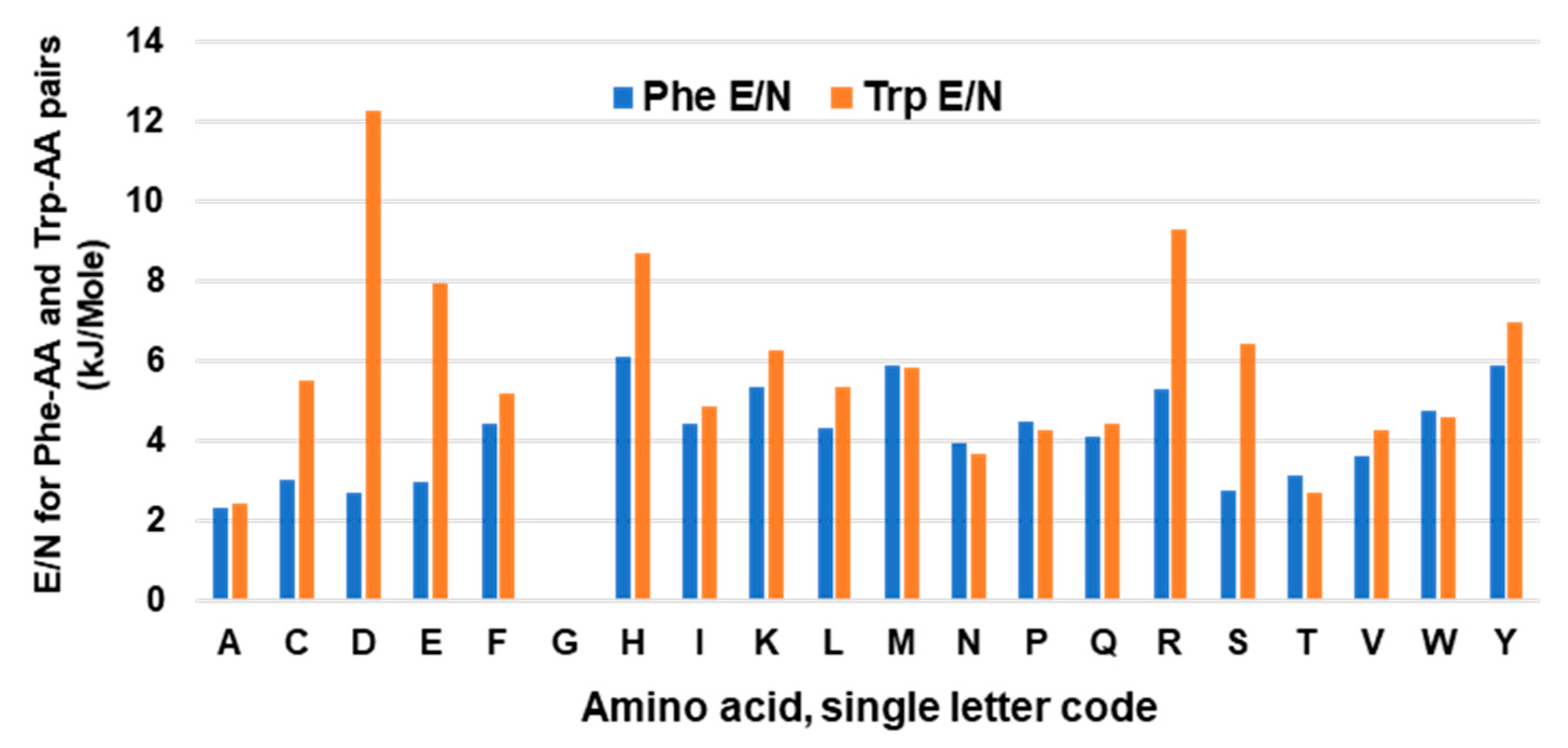
6.1. Nature of Trp Side Chain Interactions
6.2. The Energy Landscape of Trp Side Chain Interactions
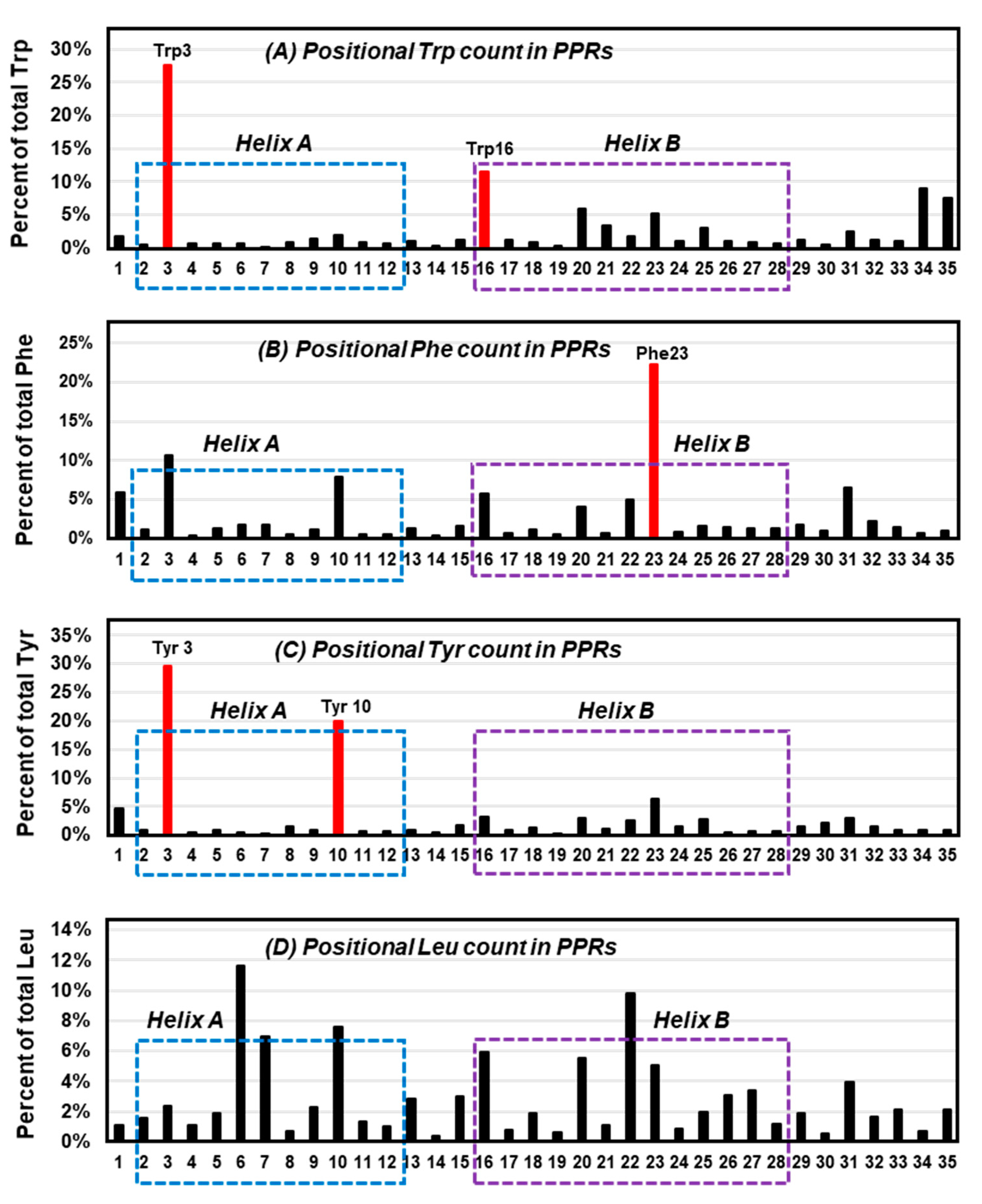
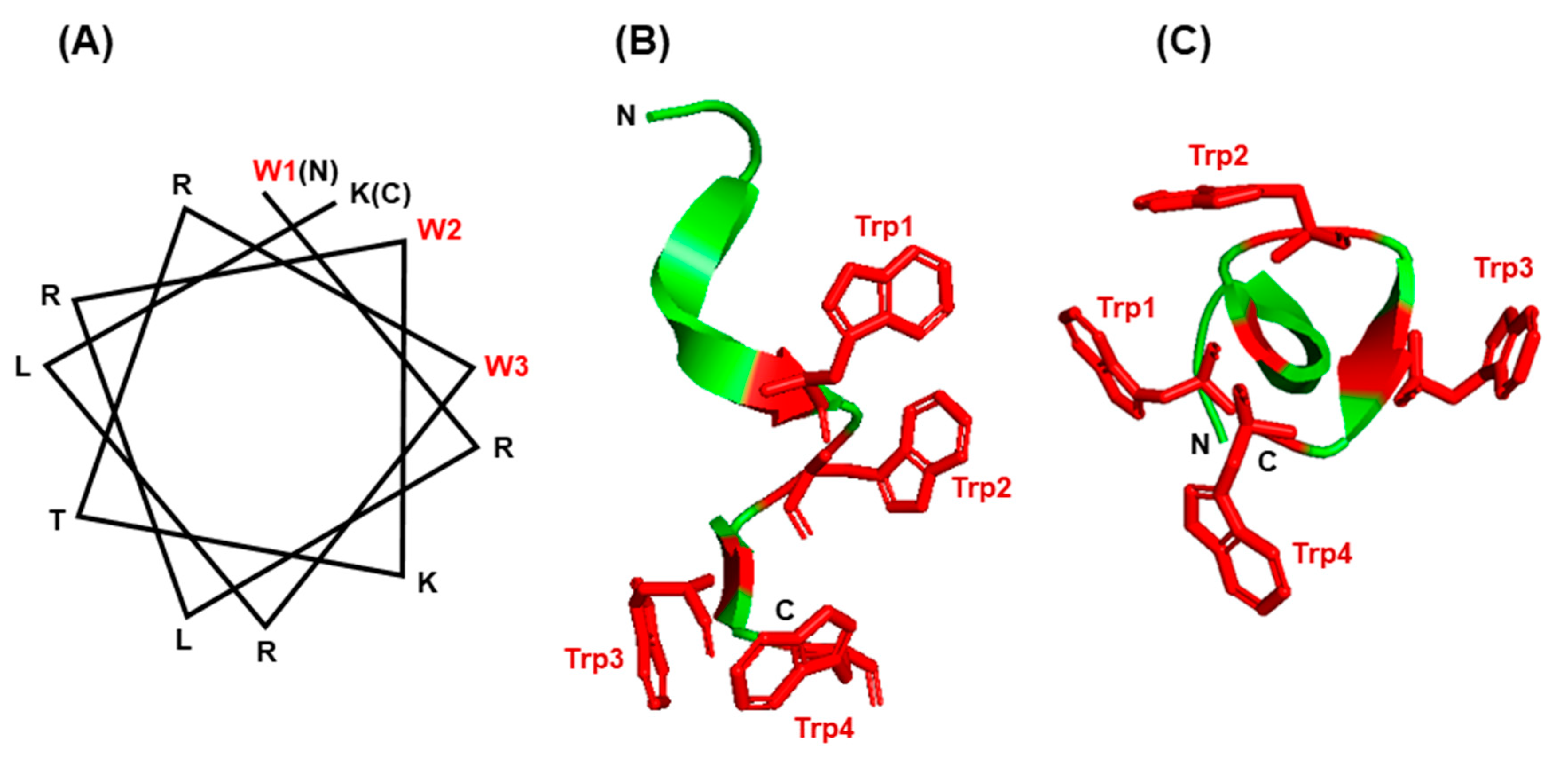
6.3. Trp in Membrane Proteins and Antimicrobial Peptides
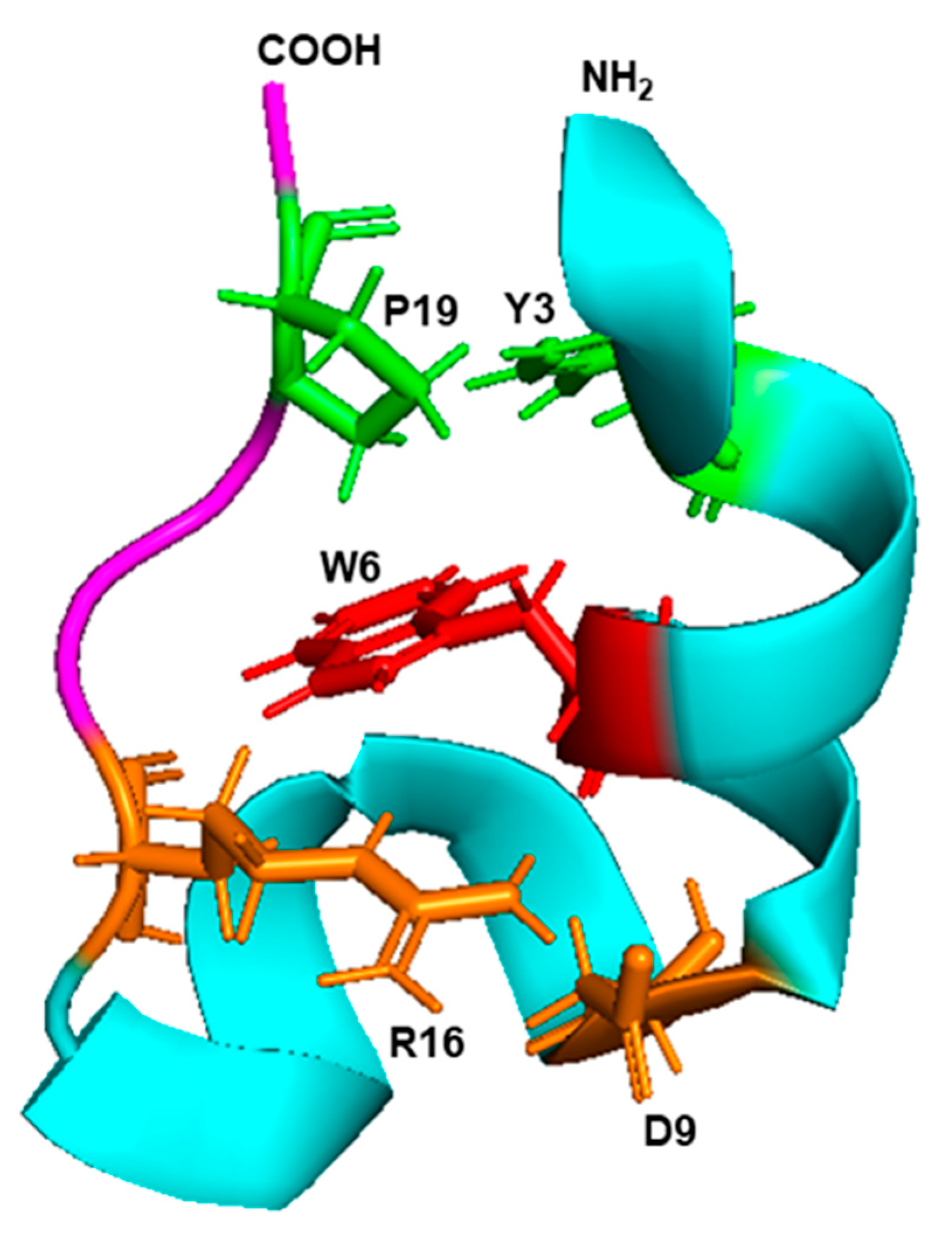
6.4. The Trp-Cage Family of Fast-Folding Peptides
7. Intersection between Trp Degradation and Immunity
8. Summary and Conclusions
Funding
Conflicts of Interest
Abbreviations
| AAAD | Aromatic amino acid decarboxylase |
| AMP | Antimicrobial peptide |
| CNS | Central nervous system |
| CTQ | Cysteine tryptophylquinone |
| 5-HT | 5-hydroxy-tryptamine |
| 5-HTP | 5-hydroxy-tryptophan |
| IDO | Indole 2,3-dioxygenase |
| IFN | Interferon |
| LPS | Lipopolysaccharide |
| MADH | Methylamine dehydrogenase |
| OTC | Over the counter (non-prescription) |
| PIV3 | Parainfluenza virus Type 3 |
| PPR | Pentatricopeptide repeat |
| TDO | Tryptophan 2,3-dehydrogenase |
| TH | Tyrosine hydroxylase |
| TPH | Tryptophan hydroxylase |
| TPR | Tetratricopeptide |
| TTQ | Tryptophan tryptophylquinone |
References
- Wu, G.; Bazer, F.W.; Dai, Z.; Li, D.; Wang, J.; Wu, Z. Amino acid nutrition in animals: Protein synthesis and beyond. Annu. Rev. Anim. Biosci. 2014, 2, 387–417. [Google Scholar] [CrossRef] [PubMed]
- Binks, H.; Vincent, G.E.; Gupta, C.; Irwin, C.; Khalesi, S. Effects of diet on sleep: A narrative. Nutrients 2020, 12, 936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chojnacki, C.; Popławski, T.; Chojnacki, J.; Fila, M.; Konrad, P.; Blasiak, J. Tryptophan intake and metabolism in older adults with mood disorders. Nutrients 2020, 12, 3183. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, A.M.; Tanabe, A.; Iwahori, Y. A systematic review of the effect of L-tryptophan supplementation on mood and emotional functioning. J. Diet. Suppl. 2020, 1–18. [Google Scholar] [CrossRef]
- Simonson, M.; Boirie, Y.; Guillet, C. Protein, amino acids and obesity treatment. Rev. Endocr. Metab. Disord. 2020, 21, 341–353. [Google Scholar] [CrossRef]
- Taleb, S. Tryptophan dietary impacts gut barrier and metabolic diseases. Front. Immunol. 2019, 10, 2113. [Google Scholar] [CrossRef]
- Khorana, H.G.; Buchi, H.; Ghosh, H.; Gupta, N.; Jacob, T.M.; Kossel, H.; Morgan, R.; Narang, S.A.; Ohtsuka, E.; Wells, R.D. Polynucleotide synthesis and the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 39–49. [Google Scholar] [CrossRef]
- Nirenberg, M.; Caskey, T.; Marshall, R.; Brimacombe, R.; Kellogg, D.; Doctor, B.; Hatfield, D.; Levin, J.; Rottman, F.; Pestka, S.; et al. The RNA code and protein synthesis. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 11–24. [Google Scholar] [CrossRef]
- Gardini, S.; Cheli, S.; Baroni, S.; Di Lascio, G.; Mangiavacchi, G.; Micheletti, N.; Monaco, C.L.; Savini, L.; Alocci, D.; Mangani, S.; et al. On Nature’s strategy for assigning genetic code multiplicity. PLoS ONE 2016, 11, e0148174. [Google Scholar] [CrossRef]
- Crick, F.H.C. Codon-anticodon pairing: The wobble hypothesis. J. Mol. Biol. 1966, 19, 548–555. [Google Scholar] [CrossRef]
- Limmer, S.; Reif, B.; Ott, G.; Arnold, L.; Sprinzl, M. NMR evidence for helix geometry modifications by a G-U wobble base pair in the acceptor arm of E. coli tRNA(Ala). FEBS Lett. 1996, 385, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Murphy, I.V.; Frank, V.; Ramakrishnan, V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat. Struct. Mol. Biol. 2004, 11, 1251–1252. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, K.; Izawa, T.; Inada, T. Recent progress on the molecular mechanism of quality controls induced by ribosome stalling. Front. Genet. 2018, 9, 743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bitran, A.; Jacobs, W.M.; Zhai, X.; Shakhnovich, E. Cotranslational folding allows misfolding-prone proteins to circumvent deep kinetic traps. Proc. Natl. Acad. Sci. USA 2020, 117, 1485–1495. [Google Scholar] [CrossRef]
- Collart, M.A.; Weiss, B. Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res. 2020, 48, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Braus, G.H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: A model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 1991, 55, 349–370. [Google Scholar] [CrossRef]
- Radwanski, E.R.; Last, R.L. Tryptophan biosynthesis and metabolism: Biochemical and molecular genetics. Plant Cell 1995, 7, 921–934. [Google Scholar] [CrossRef] [Green Version]
- Miles, E.W. The tryptophan synthase α2β2 complex: A model for substrate channeling, allosteric communication, and pyridoxal phosphate catalysis. J. Biol. Chem. 2013, 288, 10084–10091. [Google Scholar] [CrossRef] [Green Version]
- Akashi, H.; Gojobori, T. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2002, 99, 3695–3700. [Google Scholar] [CrossRef] [Green Version]
- Le Floc’h, N.; Otten, W.; Merlot, E. Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids 2011, 41, 1195–1205. [Google Scholar] [CrossRef]
- Rabbani, M.A.G.; Barik, S. 5-Hydroxytryptophan, a major product of tryptophan degradation, is essential for optimal replication of human parainfluenza virus. Virology 2017, 503, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Walther, D.J.; Peter, J.-U.; Winter, S.; Höltje, M.; Paulmann, N.; Grohmann, M.; Vowinckel, J.; Alamo-Bethencourt, V.; Wilhelm, C.S.; Ahnert-Hilger, G.; et al. Serotonylation of small GTPases is a signal transduction pathway that triggers platelet α-granule release. Cell 2003, 115, 851–862. [Google Scholar] [CrossRef] [Green Version]
- Roberts, K.M.; Fitzpatrick, P.F. Mechanisms of tryptophan and tyrosine hydroxylase. IUBMB Life 2013, 65, 350–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, R.; Urade, U.; Tokuda, M.; Hayaishi, O. Induction of indoleamine 2,3-dioxygenase in mouse lung during virus infection. Proc. Natl. Acad. Sci. USA 1979, 76, 4084–4086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munn, D.H.; Mellor, A.L. Indoleamine 2,3 dioxygenase and metabolic control of immune responses. Trends Immunol. 2013, 34, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Ball, H.J.; Sanchez-Perez, A.; Weiser, S.; Austin, C.J.D.; Astelbauer, F.; Miu, J.; McQuillan, J.A.; Stocker, R.; Jermiin, L.S.; Hunt, N.H. Characterization of an indoleamine 2,3-dioxygenase-like protein found in humans and mice. Gene 2007, 396, 203–213. [Google Scholar] [CrossRef]
- Metz, R.; Smith, C.; DuHadaway, J.B.; Chandler, P.; Baban, B.; Merlo, L.M.F.; Pigott, E.; Keough, M.P.; Rust, S.; Mellor, A.L.; et al. IDO2 is critical for IDO1-mediated T-cell regulation and exerts a non-redundant function in inflammation. Int. Immunol. 2014, 26, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Merlo, L.M.F.; DuHadaway, J.B.; Montgomery, J.D.; Peng, W.-D.; Murray, P.J.; Prendergast, G.C.; Caton, A.J.; Muller, A.J.; Mandik-Nayak, M. Differential roles of IDO1 and IDO2 in T and B cell inflammatory immune responses. Front. Immunol. 2020, 11, 1861. [Google Scholar] [CrossRef]
- Monti, J.M. Serotonin control of sleep-wake behavior. Sleep. Med. Rev. 2011, 15, 269–281. [Google Scholar] [CrossRef]
- Berger, M.; Gray, J.A.; Roth, B.L. The expanded biology of serotonin. Ann. Rev. Med. 2009, 60, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Nichols, D.E. Psychedelics. Pharmacol. Rev. 2016, 68, 264–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gershon, M.D. Review article: Serotonin receptors and transporters–oles in normal and abnormal gastrointestinal motility. Aliment. Pharmacol. Ther. 2004, 20 (Suppl. 7), 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hoyer, D. Targeting the 5-HT system: Potential side effects. Neuropharmacology 2020, 179, 108233. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.K.; Clarke, D.D.; Waelsch, H. An enzymically catalyzed incorporation of amines into proteins. Biochim. Biophys. Acta 1957, 25, 451–452. [Google Scholar] [CrossRef]
- Walther, D.J.; Peter, J.U.; Bashammakh, S.; Hörtnagl, H.; Voits, M.; Fink, H.; Bader, M. Synthesis of serotonin by a second tryptophan hydroxylase isoform. Science 2003, 299, 76. [Google Scholar] [CrossRef]
- Bader, M. Serotonylation: Serotonin signaling and epigenetics. Front. Mol. Neurosci. 2019, 12, 288. [Google Scholar] [CrossRef] [Green Version]
- Javelle, F.; Lampit, A.; Bloch, W.; Häussermann, P.; Johnson, S.L.; Zimmer, P. Effects of 5-hydroxytryptophan on distinct types of depression: A systematic review and meta-analysis. Nutr. Rev. 2020, 78, 77–88. [Google Scholar] [CrossRef]
- Shaw, K.A.; Turner, J.; Del Mar, C. Tryptophan and 5-Hydroxytryptophan for depression. Cochrane Database Syst. Rev. 2002, 1, CD003198. [Google Scholar] [CrossRef] [Green Version]
- Claustrat, B.; Leston, J. Melatonin: Physiological effects in humans. Neurochirurgie 2015, 61, 77–84. [Google Scholar] [CrossRef]
- Rios, E.R.V.; Venâncio, E.T.; Rocha, N.F.M.; Woods, D.J.; Vasconcelos, S.; Macedo, D.; de Sousa, F.C.F.; Fonteles, M.M.F. Melatonin: Pharmacological aspects and clinical trends. Int. J. Neurosci. 2010, 120, 583–590. [Google Scholar] [CrossRef]
- Baglioni, C.; Bostanova, Z.; Bacaro, V.; Benz, F.; Hertenstein, E.; Spiegelhalder, K.; Rücker, G.; Frase, L.; Riemann, D.; Feige, B. A systematic review and network meta-analysis of randomized controlled trials evaluating the evidence base of melatonin, light exposure, exercise, and complementary and alternative medicine for patients with insomnia disorder. J. Clin. Med. 2020, 9, 1949. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, V.; Spence, D.W.; Pandi-Perumal, S.R.; Trakht, I.; Cardinali, D.P. Jet lag: Therapeutic use of melatonin and possible application of melatonin analogs. Travel Med. Infect. Dis. 2008, 6, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Pandi-Perumal, S.R.; Srinivasan, V.; Maestroni, G.J.; Cardinali, D.P.; Poeggeler, B.; Hardeland, R. Melatonin: Nature’s most versatile biological signal? FEBS J. 2006, 273, 2813–2838. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Gao, R.; Wang, Z.; Wang, X.; Fang, Y.; Gao, J.; Reiter, R.J.; Wang, J. Melatonin: A potential therapeutic option for breast cancer. Trends Endocrinol. Metab. 2020, 31, 859–871. [Google Scholar] [CrossRef]
- Bondy, S.C.; Campbell, A. Melatonin and regulation of immune function: Impact on numerous diseases. Curr. Aging Sci. 2020. Online ahead of print. [Google Scholar] [CrossRef]
- Schwarcz, R.; Bruno, J.P.; Muchowski, P.J.; Wu, H.Q. Kynurenines in the mammalian brain: When physiology meets pathology. Nat. Rev. Neurosci. 2012, 13, 465–477. [Google Scholar] [CrossRef]
- Proietti, E.; Rossini, S.; Grohmann, U.; Mondanelli, G. Polyamines and kynurenines at the intersection of immune modulation. Trends Immunol. 2020, 41, 1037–1050. [Google Scholar] [CrossRef]
- Biernacki, T.; Sandi, D.; Bencsik, K.; Vécsei, L. Kynurenines in the pathogenesis of multiple sclerosis: Therapeutic perspectives. Cells 2020, 9, 1564. [Google Scholar] [CrossRef]
- Huang, Y.S.; Ogbechi, J.; Clanchy, F.I.; Williams, R.O.; Stone, T.W. IDO and kynurenine metabolites in peripheral and CNS disorders. Front. Immunol. 2020, 11, 388. [Google Scholar] [CrossRef] [Green Version]
- Zhai, L.; Bell, A.; Ladomersky, E.; Lauing, K.L.; Bollu, L.; Sosman, J.A.; Zhang, B.; Wu, J.D.; Miller, S.D.; Meeks, J.J.; et al. Immunosuppressive IDO in cancer: Mechanisms of action, animal models, and targeting strategies. Front. Immunol. 2020, 11, 1185. [Google Scholar] [CrossRef]
- Lee, J.H.; Wood, T.K.; Lee, J. Roles of indole as an interspecies and interkingdom signaling molecule. Trends Microbiol. 2015, 23, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Sardar, P.; Kempken, F. Characterization of indole-3-pyruvic acid pathway-mediated biosynthesis of auxin in Neurospora crassa. PLoS ONE 2018, 13, e0192293. [Google Scholar] [CrossRef] [PubMed]
- González, B.; Vázquez, J.; Morcillo-Parra, M.Á.; Mas, A.; Torija, M.J.; Beltran, G. The production of aromatic alcohols in non-Saccharomyces wine yeast is modulated by nutrient availability. Food Microbiol. 2018, 74, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Cornford, E.M.; Bocash, W.D.; Braun, L.D.; Crane, P.D.; Oldendorf, W.H.; MacInnis, A.J. Rapid distribution of tryptophol (3-indole ethanol) to the brain and other tissues. J. Clin. Investig. 1979, 63, 1241–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldstein, A.; Chang, F.H.; Kucharski, J.M. Tryptophol, 5-hydroxytryptophol and 5-methoxytryptophol induced sleep in mice. Life Sci. 1970, 9, 323–329. [Google Scholar] [CrossRef]
- Laćan, G.; Magnus, V.; Jeričević, B.; Kunst, L.; Iskrić, S. Formation of tryptophol galactoside and an unknown tryptophol ester in Euglena gracilis. Plant Physiol. 1984, 76, 889–893. [Google Scholar] [CrossRef] [Green Version]
- Hofsteenge, J.; Müller, D.R.; de Beer, T.; Löffler, A.; Richter, W.J.; Vliegenthart, J.F. New type of linkage between a carbohydrate and a protein: C-glycosylation of a specific tryptophan residue in human RNase Us. Biochemistry 1994, 33, 13524–13530. [Google Scholar] [CrossRef] [Green Version]
- Gutsche, B.; Grun, C.; Scheutzow, D.; Herderich, M. Tryptophan glycoconjugates in food and human urine. Biochem. J. 1999, 343, 11–19. [Google Scholar] [CrossRef]
- Palmieri, A.; Petrini, M. Tryptophol and derivatives: Natural occurrence and applications to the synthesis of bioactive compounds. Nat. Prod. Rep. 2019, 36, 490–530. [Google Scholar] [CrossRef]
- Ray, P.D.; Foster, D.O.; Lardy, H.A. Paths of carbon in gluconeogenesis and lipogenesis. IV. Inhibition by L-tryptophan of hepatic gluconeogenesis at the level of phosphoenolpyruvate formation. J. Biol. Chem. 1966, 241, 3904–3908. [Google Scholar]
- Smith, S.A.; Elliott, K.R.F.; Pogson, C.I. Differential effects of tryptophan on glucose synthesis in rats and guinea pigs. Biochem. J. 1978, 176, 817–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badawy, A.A. Kynurenine pathway of tryptophan metabolism: Regulatory and functional aspects. Int. J. Tryptophan Res. 2017, 10, 1178646917691938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, V.L.; Liu, A. Tryptophan tryptophylquinone biosynthesis: A radical approach to posttranslational modification. Biochim. Biophys. Acta 2012, 1824, 1299–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, V.L. Protein-derived cofactors revisited: Empowering amino acid residues with new functions. Biochemistry 2018, 57, 3115–3125. [Google Scholar] [CrossRef]
- McIntire, W.S.; Wemmer, D.E.; Chistoserdov, A.; Lidstrom, M.E. A new cofactor in a prokaryotic enzyme: Tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science 1991, 252, 817–824. [Google Scholar] [CrossRef]
- Davidson, V.L.; Wilmot, C.M. Posttranslational biosynthesis of the protein-derived cofactor tryptophan tryptophylquinone. Annu. Rev. Biochem. 2013, 82, 531–550. [Google Scholar] [CrossRef] [Green Version]
- Davidson, V.L. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv. Protein. Chem. 2001, 58, 95–140. [Google Scholar] [CrossRef]
- Datta, S.; Mori, Y.; Takagi, K.; Kawaguchi, K.; Chen, Z.W.; Okajima, T.; Kuroda, S.; Ikeda, T.; Kano, K.; Tanizawa, K.; et al. Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc. Natl. Acad. Sci. USA 2001, 98, 14268–14273. [Google Scholar] [CrossRef] [Green Version]
- Campillo-Brocal, J.C.; Chacon-Verdu, M.D.; Lucas-Elio, P.; Sanchez-Amat, A. Distribution in microbial genomes of genes similar to lodA and goxA which encode a novel family of quinoproteins with amino acid oxidase activity. BMC Genom. 2015, 16, 231. [Google Scholar] [CrossRef] [Green Version]
- Campillo-Brocal, J.C.; Lucas-Elio, P.; Sanchez-Amat, A. Distribution in different amino acid oxidases with FAD or a quinone as cofactor and their role as antimicrobial proteins in marine bacteria. Mar. Drugs 2015, 13, 7403–7418. [Google Scholar] [CrossRef] [Green Version]
- Schlamadinger, D.E.; Gable, J.E.; Kim, J.E. Hydrogen bonding and solvent polarity markers in the UV resonance Raman spectrum of tryptophan: Application to membrane proteins. J. Phys. Chem. B 2009, 113, 14769–14778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas Jr, G.J. New structural insights from Raman spectroscopy of proteins and their assemblies. Biopolymers 2002, 67, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, H. UV Raman markers for structural analysis of aromatic side chains in proteins. Anal. Sci. 2011, 27, 1077–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiler-Feilchenfeld, H.; Pullman, A.; Berthod, H.; Giessner-Prettre, C. Experimental and quantum-chemical studies of the dipole moments of quinoline and indole. J. Mol. Struct. 1970, 6, 297–304. [Google Scholar] [CrossRef]
- Galgonek, J.; Vymetal, J.; Jakubec, D.; Vondrášek, J. Amino Acid Interaction (INTAA) web server. Nucleic Acids Res. 2017, 45, W388–W392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barik, S. Protein tetratricopeptide repeat and the companion non-tetratricopeptide repeat helices: Bioinformatic analysis of interhelical interactions. Bioinform. Biol. Insights 2019, 13, 1177932219863363. [Google Scholar] [CrossRef] [PubMed]
- Barik, S. The nature and arrangement of pentatricopeptide domains and the linker sequences between them. Bioinform. Biol. Insights 2020, 14, 1177932220906434. [Google Scholar] [CrossRef] [Green Version]
- Small, I.D.; Peeters, N. The PPR motif–a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000, 25, 46–47. [Google Scholar] [CrossRef]
- Barkan, A.; Small, I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant. Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Main, E.R.G.; Xiong, Y.; Cocco, M.J.; D’Andrea, L.; Regan, L. Design of stable alpha-helical arrays from an idealized TPR motif. Structure 2003, 11, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Gallivan, J.P.; Dougherty, D.A. Cation-pi interactions in structural biology. Proc. Natl. Acad. Sci. USA 1999, 96, 9459–9464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, C.A.; Shi, Z.; Kallenbach, N.R. Polar interactions with aromatic side chains in alpha-helical peptides: Ch...O H-bonding and cation-pi interactions. J. Am. Chem. Soc. 2001, 123, 6451–6452. [Google Scholar] [CrossRef] [PubMed]
- Hait, S.; Mallik, S.; Basu, S.; Kundu, S. Finding the generalized molecular principles of protein thermal stability. Proteins 2020, 88, 788–808. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayasinghe, S.; Hristova, K.; White, S.H. MPtopo: A database of membrane protein topology. Protein Sci. 2001, 10, 455–458. [Google Scholar] [CrossRef] [Green Version]
- Granseth, E.; von Heijne, G.; Elofsson, A. A study of the membrane-water interface region of membrane proteins. J. Mol. Biol. 2005, 346, 377–385. [Google Scholar] [CrossRef]
- Sanchez, K.M.; Gable, J.E.; Schlamadinger, D.E.; Kim, J.E. Effects of tryptophan microenvironment, soluble domain, and vesicle size on the thermodynamics of membrane protein folding: Lessons from the transmembrane protein OmpA. Biochemistry 2008, 47, 12844–12852. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, K.M.; Kang, G.; Wu, B.; Kim, J.E. Tryptophan-lipid interactions in membrane protein folding probed by ultraviolet resonance Raman and fluorescence spectroscopy. Biophys. J. 2011, 100, 2121–2130. [Google Scholar] [CrossRef] [Green Version]
- Persson, S.; Killian, J.A.; Lindblom, G. Molecular ordering of interfacially localized tryptophan analogs in ester- and ether-lipid bilayers studied by H-2-NMR. Biophys. J. 1998, 75, 1365–1371. [Google Scholar] [CrossRef] [Green Version]
- van der Wel, P.C.A.; Reed, N.D.; Koeppe, R.E. Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry 2007, 46, 7514–7524. [Google Scholar] [CrossRef] [Green Version]
- Shafaat, H.S.; Sanchez, K.M.; Kim, J.E. Ultraviolet resonance Raman spectroscopy of a membrane-bound beta-sheet peptide as a model for membrane protein folding. J. Raman Spectrosc. 2008, 40, 1060–1064. [Google Scholar] [CrossRef]
- Sun, H.; Greathouse, D.V.; Koeppe, R.E. The preference of tryptophan for membrane interfaces: Insights from N-methylation of tryptophans in gramicidin channels. J. Biol. Chem. 2008, 283, 22233–22243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haney, E.F.; Straus, S.K.; Hancock, R.E.W. Reassessing the host defense peptide landscape. Front. Chem. 2019, 7, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, H.; Park, S.; Jimenez, R.H.F.; Rinehard, D.; Tamm, L.K. Role of aromatic side chains in the folding and thermodynamic stability of integral membrane proteins. J. Am. Chem. Soc. 2007, 129, 8320–8327. [Google Scholar] [CrossRef]
- Bhattacharjya, S.; Straus, S.K. Design, engineering and discovery of novel α-helical and β-boomerang antimicrobial peptides against drug resistant bacteria. Int. J. Mol. Sci. 2020, 21, 5773. [Google Scholar] [CrossRef]
- Necelis, M.R.; Santiago-Ortiz, L.E.; Caputo, G.A. Investigation of the role of aromatic residues in the antimicrobial peptide BuCATHL4B. Protein Pept. Lett. 2020. Online ahead of print. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, J.; Tong, Z.; Jia, Y.; Yang, K.; Wang, Z. Potent broad-spectrum antibacterial activity of amphiphilic peptides against multidrug-resistant bacteria. Microorganisms 2020, 8, 1398. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, D.A.; Chattopadhyay, A. The gramicidin ion channel: A model membrane protein. Biochim. Biophys. Acta. 2007, 1768, 2011–2025. [Google Scholar] [CrossRef] [Green Version]
- Becker, M.D.; Greathouse, D.V.; Koeppe, R.E.; Andersen, O.S. Amino acid sequence modulation of gramicidin channel function: Effects of tryptophan-to-phenylalanine substitutions on the single-channel conductance and duration. Biochemistry 1991, 30, 8830–8839. [Google Scholar] [CrossRef]
- Salom, D.; Pérez-Payá, E.; Pascal, J.; Abad, C. Environment- and sequence-dependent modulation of the double-stranded to single-stranded conformational transition of gramicidin A in membranes. Biochemistry 1998, 37, 14279–14291. [Google Scholar] [CrossRef]
- Cotten, M.; Xu, F.; Cross, T.A. Protein stability and conformational rearrangements in lipid bilayers: Linear gramicidin, a model system. Biophys. J. 1997, 73, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Chaudhuri, A.; Haldar, S.; Sun, H.; Koeppe II, R.R.; Chattopadhyay, A. Importance of indole N-single bond-H hydrogen bonding in the organization and dynamics of gramicidin channels. Biochim. Biophys. Acta. 2014, 1838, 419–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheiner, S.; Kar, T.; Pattanayak, J. Comparison of various types of hydrogen bonds involving aromatic amino acids. J. Am. Chem. Soc. 2002, 124, 13257–13264. [Google Scholar] [CrossRef] [PubMed]
- Neidigh, J.W.; Fesinmeyer, R.M.; Prickett, K.S.; Andersen, N.H. Exendin-4 and glucagon-like-peptide-1: NMR structural comparisons in the solution and micelle-associated states. Biochemistry 2001, 40, 13188–13200. [Google Scholar] [CrossRef]
- Neidigh, J.W.; Fesinmeyer, R.M.; Andersen, N.H. Designing a 20-residue protein. Nat. Struct. Biol. 2002, 9, 425–430. [Google Scholar] [CrossRef]
- Qiu, L.; Pabit, S.A.; Roitberg, A.E.; Hagen, S.J. Smaller and faster: The 20-residue Trp-cage protein folds in 4 micros. J. Am. Chem. Soc. 2002, 124, 12952–12953. [Google Scholar] [CrossRef]
- Byrne, A.; Williams, D.V.; Barua, B.; Hagen, S.J.; Kier, B.L.; Andersen, N.H. Folding dynamics and pathways of the trp-cage miniproteins. Biochemistry 2014, 53, 6011–6021. [Google Scholar] [CrossRef]
- Barua, B.; Lin, J.C.; Williams, V.D.; Kummler, P.; Neidigh, J.W.; Andersen, N.H. The Trp-cage: Optimizing the stability of a globular miniprotein. Protein Eng. Des. Sel. 2008, 21, 171–185. [Google Scholar] [CrossRef] [Green Version]
- Sobash, P.T.; Kolhe, R.; Karim, N.A.; Guddati, A.K.; Jillella, A.; Kota, V. Role of indoleamine 2,3-dioxygenase in acute myeloid leukemia. Future Oncol. 2020. Online ahead of print. [Google Scholar] [CrossRef]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-de Bretagne, I.; Cannet, P.; et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Ma, N.; He, T.; Johnston, L.J.; Ma, X. Host-microbiome interactions: The aryl hydrocarbon receptor as a critical node in tryptophan metabolites to brain signaling. Gut Microbes 2020, 11, 1203–1219. [Google Scholar] [CrossRef] [PubMed]
- Hényková, E.; Vránová, H.P.; Amakorová, P.; Pospíšil, T.; Žukauskaitė, A.; Vlčková, M.; Urbánek, L.; Novák, O.; Mareš, J.; Kaňovský, P.; et al. Stable isotope dilution ultra-high performance liquid chromatography-tandem mass spectrometry quantitative profiling of tryptophan-related neuroactive substances in human serum and cerebrospinal fluid. J. Chromatogr. A 2016, 1437, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, Y.; Edelstein, M.P.; Duch, D.S. Induction of indoleamine 2,3-dioxygenase: A mechanism of the antitumor activity of interferon gamma. Proc. Natl. Acad. Sci. USA 1988, 85, 1242–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, G.S.; Taylor, M.W. Interferon gamma-resistant mutants are defective in the induction of indoleamine 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 1989, 86, 7144–7148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frumento, G.; Rotondo, R.; Tonetti, M.; Damonte, G.; Benatti, U.; Ferrara, G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 2002, 196, 459–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwu, P.; Du, M.X.; Lapointe, R.; Do, M.; Taylor, M.W.; Young, H.A. Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J. Immunol. 2000, 164, 3596–3599. [Google Scholar] [CrossRef]
- Munn, D.H.; Sharma, M.D.; Lee, J.R.; Jhaver, K.J.; Johnson, T.S.; Keskin, D.B.; Marshall, B.; Chandler, P.; Antonia, S.J.; Burgess, R.; et al. Potential regulatory function of human dendritic cells expressing indoleamine 2,3-dioxygenase. Science 2002, 297, 1867–1870. [Google Scholar] [CrossRef]
- Orabona, C.; Grohmann, U. Indoleamine 2,3-dioxygenase and regulatory function: Tryptophan starvation and beyond. Methods Mol. Biol. 2011, 677, 269–280. [Google Scholar] [CrossRef]
- Dai, W.; Pan, H.; Kwok, O.; Dubey, J.P. Human indoleamine 2,3-dioxygenase inhibits Toxoplasma gondii growth in fibroblast cells. J. Interferon Res. 1994, 14, 313–317. [Google Scholar] [CrossRef]
- Mehraj, V.; Routy, J.P. Tryptophan catabolism in chronic viral infections: Handling uninvited guests. Int. J. Tryptophan Res. 2015, 8, 41–48. [Google Scholar] [CrossRef]
- Mellor, A.L.; Munn, D.H. Tryptophan catabolism and regulation of adaptive immunity. J. Immunol. 2003, 170, 5809–5813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, O.; Besken, K.; Oberdörfer, C.; MacKenzie, C.R.; Rüssing, D.; Däubener, W. Inhibition of human herpes simplex virus type 2 by interferon gamma and tumor necrosis factor alpha is mediated by indoleamine 2,3-dioxygenase. Microbes Infect. 2004, 6, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, L.; Klonowski, K.D.; Tompkins, S.M.; Tripp, R.A.; Mellor, A.L. Induction and role of indoleamine 2,3 dioxygenase in mouse models of influenza A virus infection. PLoS ONE 2013, 8, e66546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, R.; Zhang, J.; Jiang, D.; Cai, D.; Levy, J.M.; Cuconati, A.; Block, T.M.; Guo, J.T.; Guo, H. Indoleamine 2,3-dioxygenase mediates the antiviral effect of gamma interferon against hepatitis B virus in human hepatocyte-derived cells. J. Virol. 2011, 85, 1048–1057. [Google Scholar] [CrossRef] [Green Version]
- Obojes, K.; Andres, O.; Kim, K.S.; Däubener, W.; Schneider-Schaulies, J. Indoleamine 2,3-dioxygenase mediates cell type-specific anti-measles virus activity of gamma interferon. J. Virol. 2005, 79, 7768–7776. [Google Scholar] [CrossRef] [Green Version]
- Sage, L.K.; Fox, J.M.; Mellor, A.L.; Tompkins, S.M.; Tripp, R.A. Indoleamine 2,3-dioxygenase (IDO) activity during the primary immune response to influenza infection modifies the memory T cell response to influenza challenge. Viral Immunol. 2014, 27, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Rabbani, M.A.G.; Ribaudo, M.; Guo, J.-T.; Barik, S. Identification of interferon-stimulated gene proteins that inhibit human parainfluenza virus type 3. J. Virol. 2016, 90, 11145–11156. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.V.; Schultze, J.L. New insights into IDO biology in bacterial and viral infections. Front. Immunol. 2014, 5, 384. [Google Scholar] [CrossRef] [Green Version]
- Bitko, V.; Musiyenko, A.; Shulyayeva, O.; Barik, S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005, 11, 50–55. [Google Scholar] [CrossRef]
- Villenave, R.; Shields, M.D.; Power, U.F. Respiratory syncytial virus interaction with human airway epithelium. Trends Microbiol. 2013, 21, 238–244. [Google Scholar] [CrossRef]


| Codon | Amino Acid | Structure of Side Chain (-R) | Distinguishing Property |
|---|---|---|---|
| UGG | Trp | 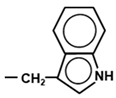 | Aromatic, hydrophobic |
| GGG | Gly | None | Smallest, flexible, no side chain |
| UCG | Ser | 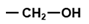 | Hydroxyl, hydrophilic |
| UGC, UGU | Cys | 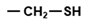 | Thiol, hydrophilic (Ser homolog) |
| UUG | Leu | 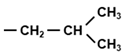 | Large, hydrophobic |
| AGG, CGG | Arg | 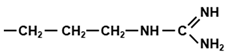 | Basic, charged, hydrophilic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barik, S. The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. Int. J. Mol. Sci. 2020, 21, 8776. https://doi.org/10.3390/ijms21228776
Barik S. The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. International Journal of Molecular Sciences. 2020; 21(22):8776. https://doi.org/10.3390/ijms21228776
Chicago/Turabian StyleBarik, Sailen. 2020. "The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins" International Journal of Molecular Sciences 21, no. 22: 8776. https://doi.org/10.3390/ijms21228776
APA StyleBarik, S. (2020). The Uniqueness of Tryptophan in Biology: Properties, Metabolism, Interactions and Localization in Proteins. International Journal of Molecular Sciences, 21(22), 8776. https://doi.org/10.3390/ijms21228776





