UGT1A1 and UGT1A9 Are Responsible for Phase II Metabolism of Tectorigenin and Irigenin In Vitro
Abstract
:1. Introduction
2. Results
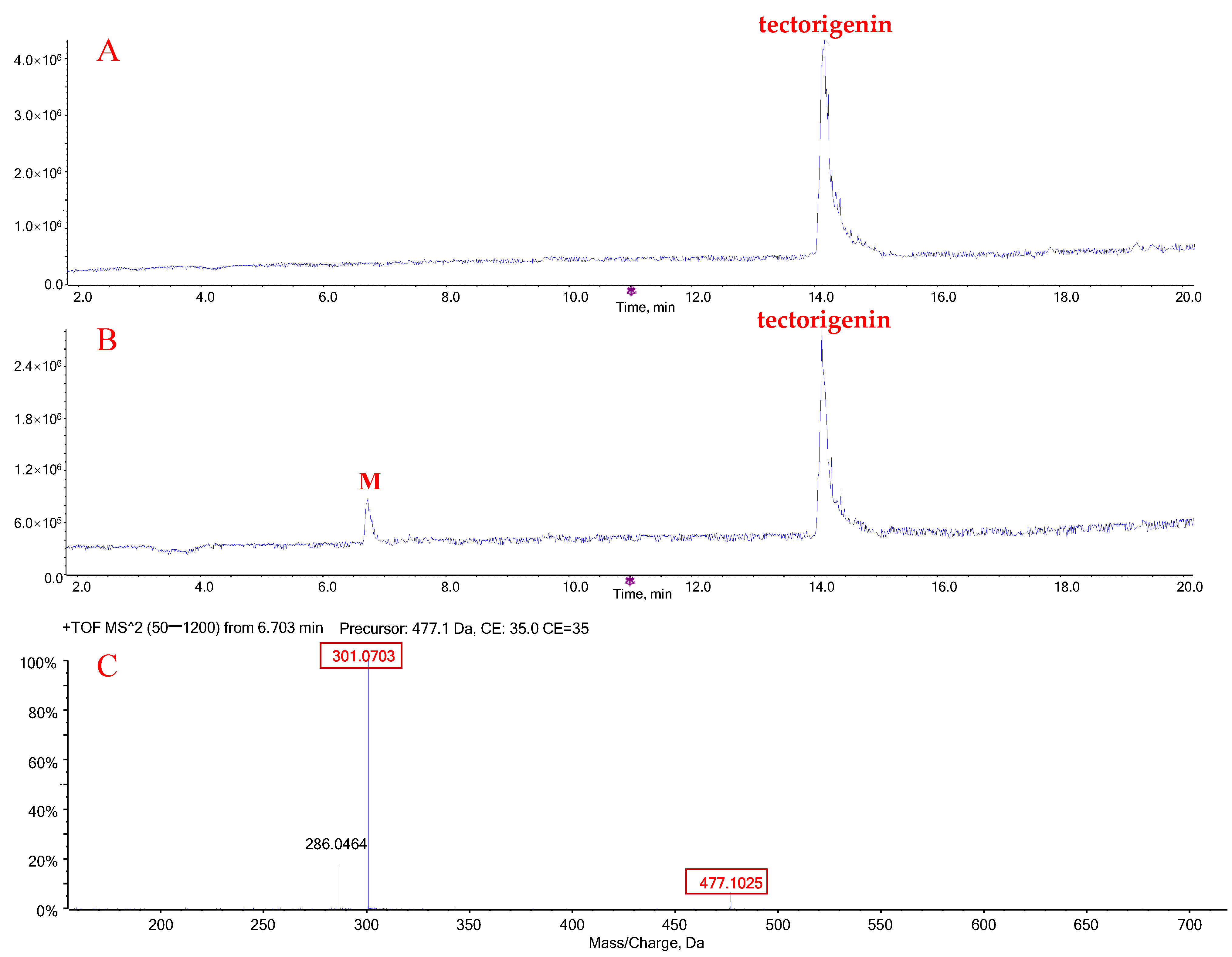
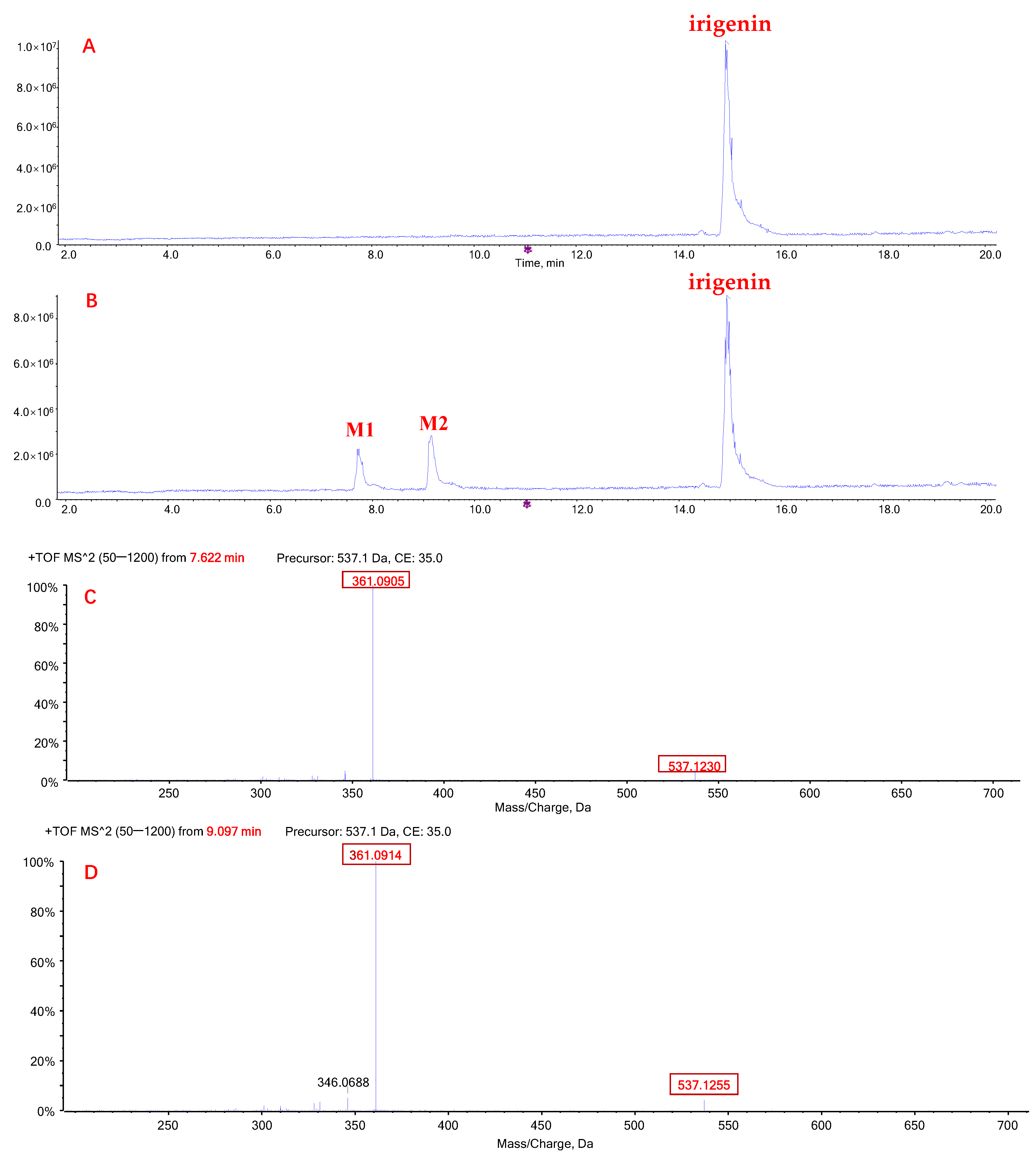
2.1. Metabolic Profiles of Tectorigenin and Irigenin
2.2. Response Factor Values
2.3. Quantification of Metabolites
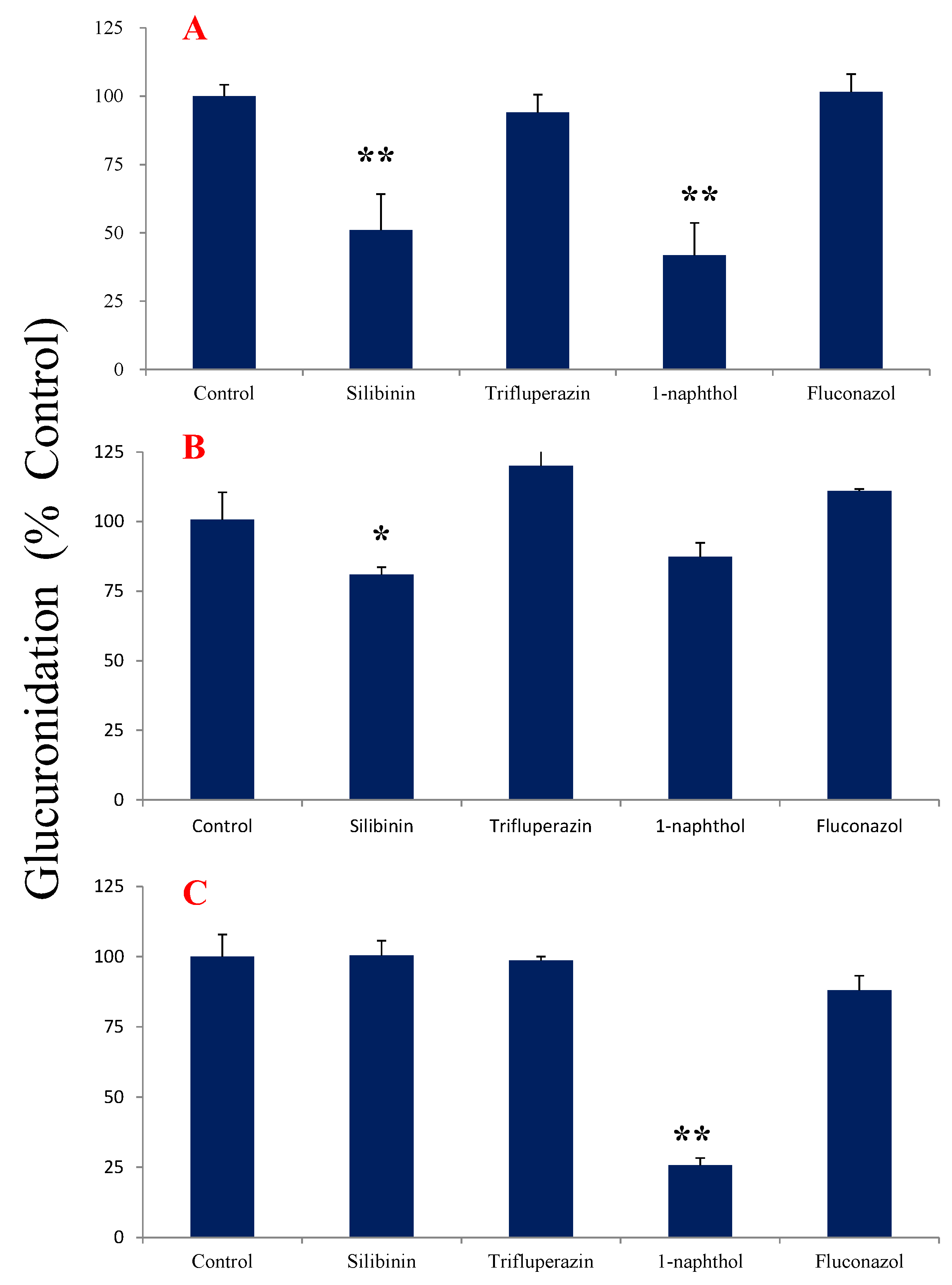
2.4. Chemical Inhibition Studies
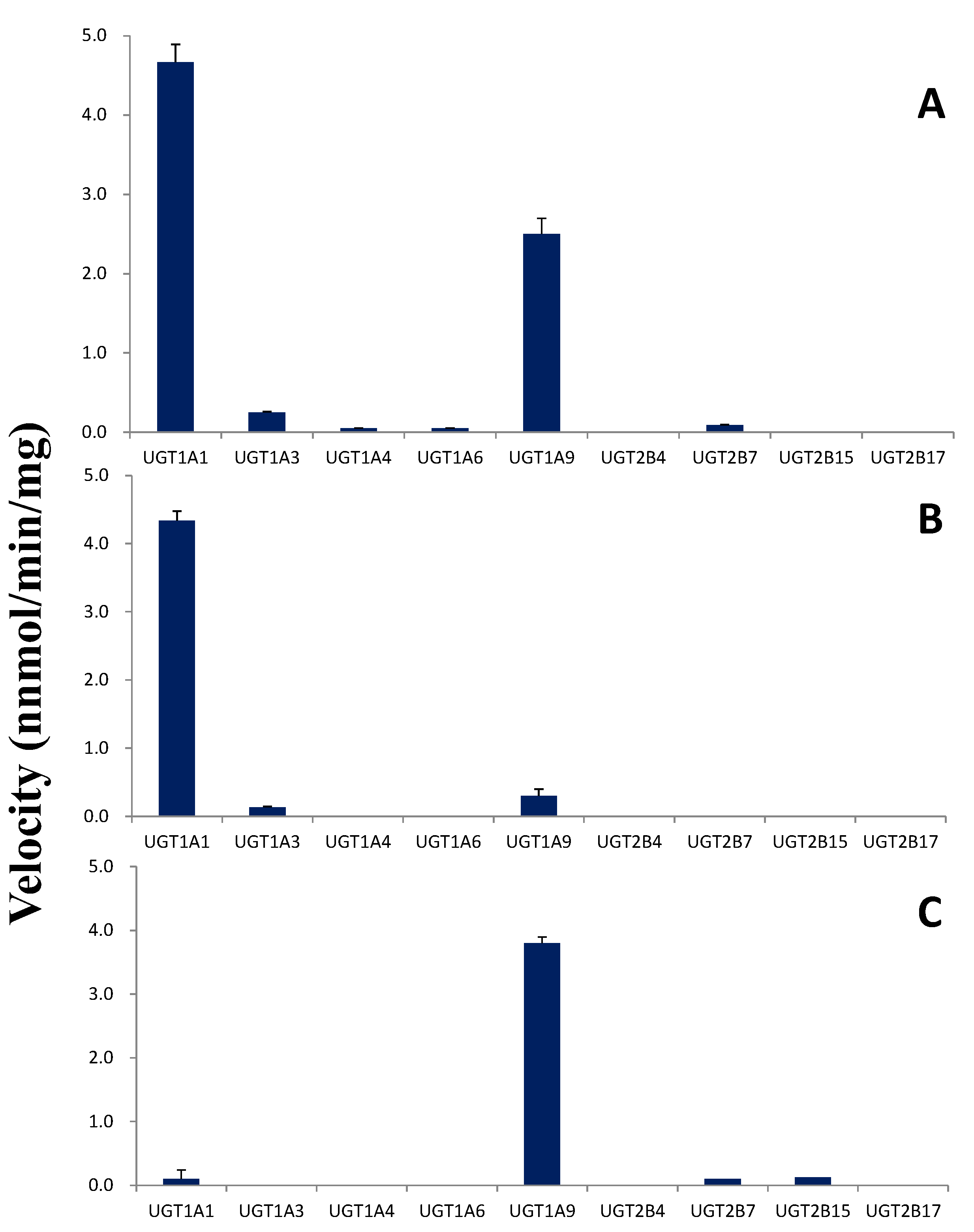
2.5. Glucuronidation by UGTs
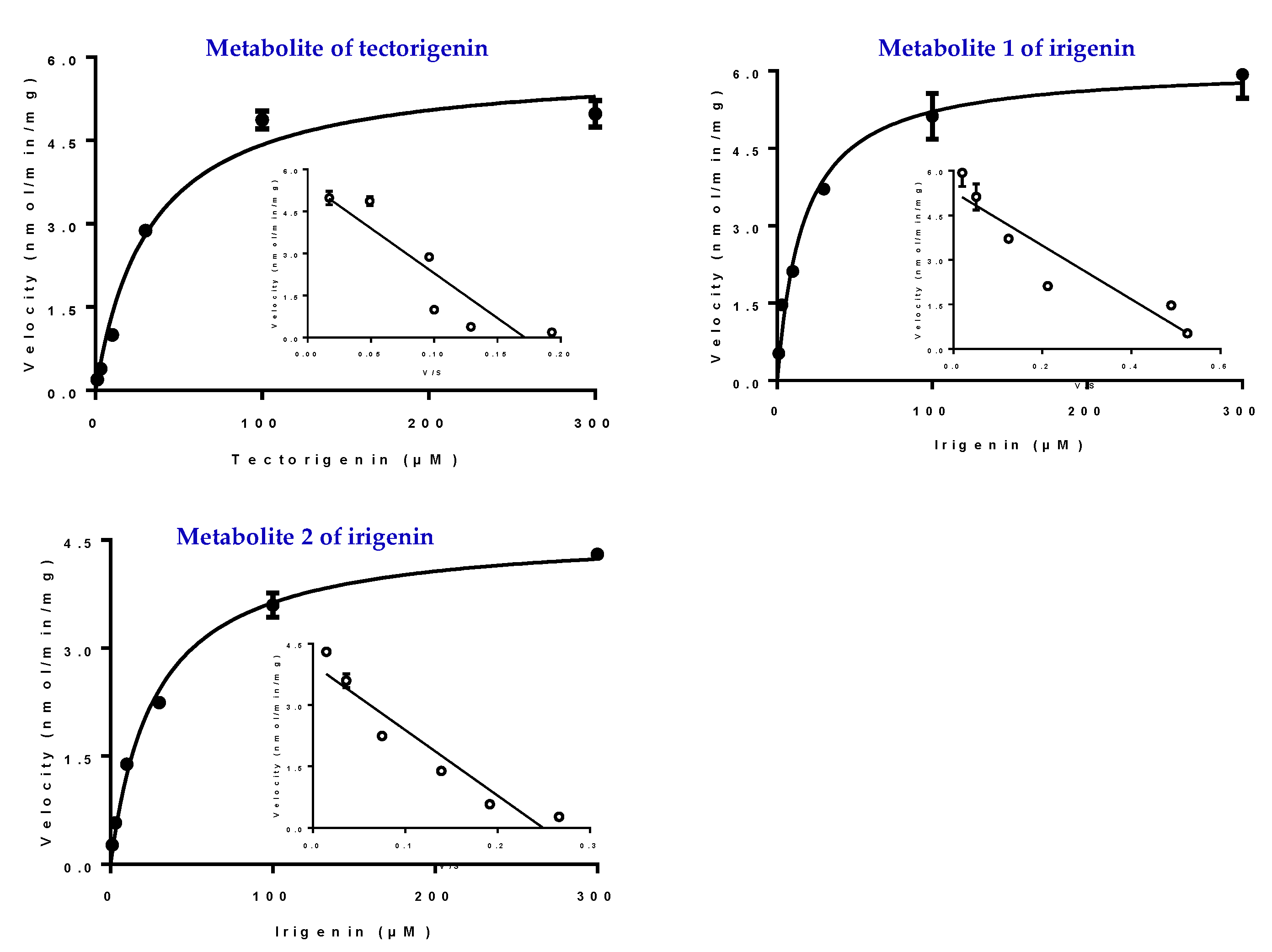
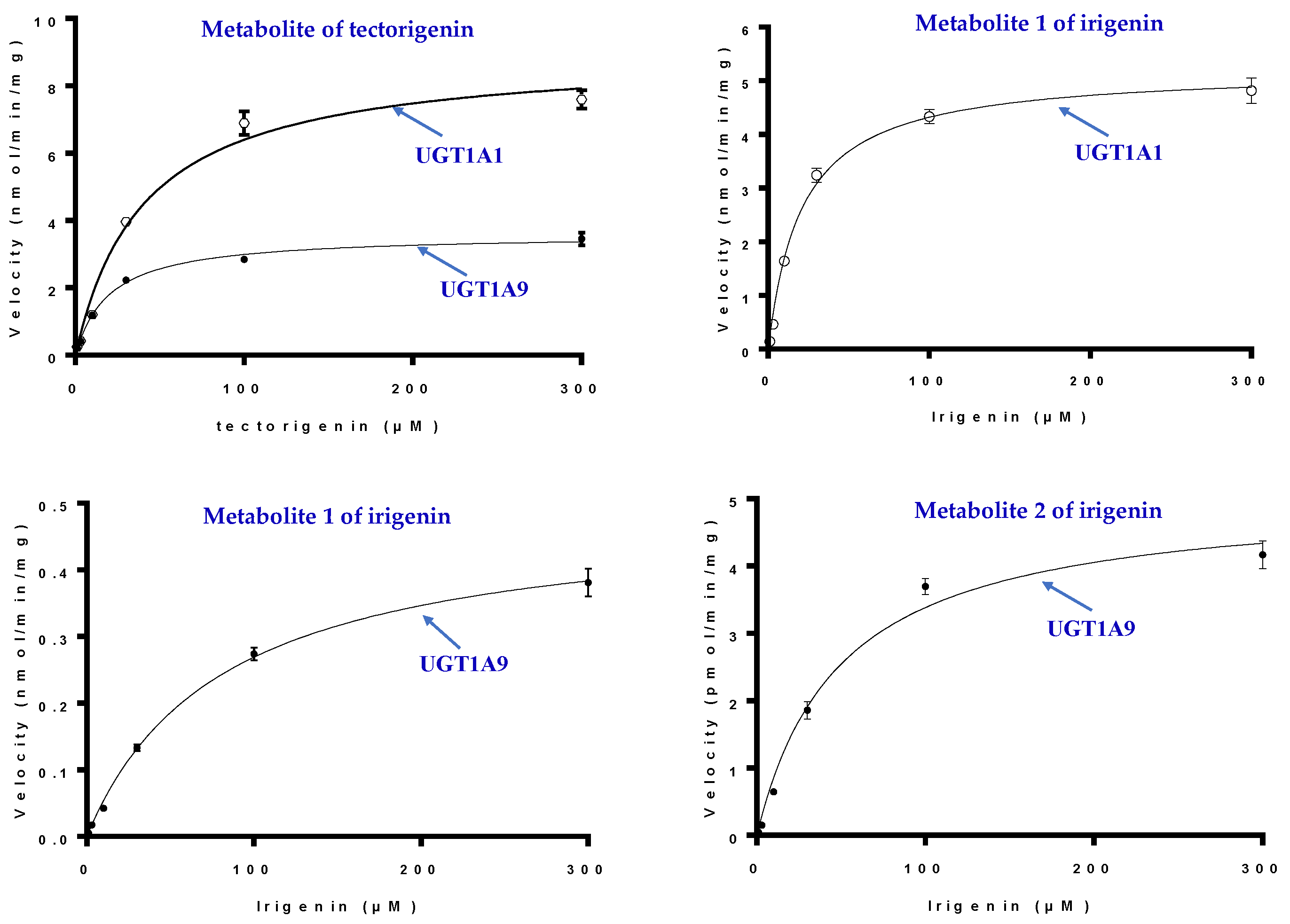
2.6. Kinetic Studies in HLMs and UGTs
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Metabolic Stability Experiments
4.3. Molecular Weight Analysis of Tectorigenin and Irigenin Glucuronides
4.4. Determination of RF Values
4.5. Quantitative Determination of Metabolites of Tectorigenin and Irigenin
4.6. Glucuronidation Kinetics of Tectorigenin and Irigenin in HLMs
4.7. Inhibition of Glucuronidation Reaction by Chemical Inhibitors
4.8. UGT Reaction Screening and Enzyme Kinetics of Tectorigenin and Irigenin
4.9. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Zhu, Y.; Pu, B.Q.; Xie, G.Y.; Tian, M.; Xu, F.Y.; Qin, M.J. Dynamic changes of flavonoids contents in the different parts of rhizome of Belamcanda chinensis during the thermal drying process. Molecules 2014, 19, 10440–10454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wei, K.; Xu, J.; Yang, D.; Zhang, C.; Wang, Z.; Li, M. Belamcanda chinensis (L.) DC-An ethnopharmacological, phytochemical and pharmacological review. J. Ethnopharmacol. 2016, 186, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, D.; Matkowski, A. Belamcandae chinensis rhizome–A review of phytochemistry and bioactivity. Fitoterapia 2015, 107, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Moqbel, S.A.A.; Xu, K.; Chen, Z.; Xu, L.; He, Y.; Wu, Z.; Ma, C.; Ran, J.; Wu, L.; Xiong, Y. Tectorigenin Alleviates Inflammation, Apoptosis, and Ossification in Rat Tendon-Derived Stem Cells via Modulating NF-Kappa B and MAPK Pathways. Front. Cell Dev. Biol. 2020, 8, 568894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Piao, M.J.; Oh, M.C.; Park, J.E.; Shilnikova, K.; Moon, Y.J.; Kim, D.H.; Jung, U.; Kim, I.G.; Hyun, J.W. Protective effect of an isoflavone, tectorigenin, against oxidative stress-induced cell death via catalase activation. J. Cancer Prev. 2016, 21, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.T.; Sohn, I.C.; Kim, D.H.; Choi, J.W.; Kwon, S.H.; Park, H.J. Hypoglycemic and hypolipidemic effects of tectorigenin and kaikasaponin III in the streptozotocin-lnduced diabetic rat and their antioxidant activity in vitro. Arch. Pharm. Res. 2000, 23, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.H.; Liu, J.P.; Qu, R.; Ma, S.P. Tectorigenin inhibits the inflammation of LPS-induced acute lung injury in mice. Chin. J. Nat. Med. 2014, 12, 841–846. [Google Scholar] [CrossRef]
- Liu, E.Y.; Zheng, Z.X.; Zheng, B.Z.; Xia, Y.; Guo, M.S.; Dong, T.T.; Tsim, K.W.K. Tectorigenin, an isoflavone aglycone from the rhizome of Belamcanda chinensis, induces neuronal expression of erythropoietin via accumulation of hypoxia-inducible factor-1α. Phytother. Res. 2020, 34, 1329–1337. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, Y.; Fan, L.; Xu, K.; Ji, F.; Xie, Z.; Ouyang, X.; Wu, D.; Li, L. Tectorigenin protects against experimental fulminant hepatic failure by regulating the TLR4/mitogen-activated protein kinase and TLR4/nuclear factor-κB pathways and autophagy. Phytother. Res. 2019, 33, 1055–1064. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Zhu, B.; Fan, J.; Hu, Q.; Wang, L. Tectorigenin protects against unilateral ureteral obstruction by inhibiting Smad3-mediated ferroptosis and fibrosis. Phytother. Res. 2022, 36, 475–487. [Google Scholar] [CrossRef]
- Guo, L.; Zheng, X.; Wang, E.; Jia, X.; Wang, G.; Wen, J. Irigenin treatment alleviates doxorubicin (DOX)-induced cardiotoxicity by suppressing apoptosis, inflammation and oxidative stress via the increase of miR-425. Biomed. Pharmacother. 2020, 125, 109784. [Google Scholar] [CrossRef] [PubMed]
- Amin, A.; Chikan, N.A.; Mokhdomi, T.A.; Bukhari, S.; Koul, A.M.; Shah, B.A.; Gharemirshamlu, F.R.; Wafai, A.H.; Qadri, A.; Qadri, R.A. Irigenin, a novel lead from Western Himalayan chemiome inhibits Fibronectin-Extra Domain A induced metastasis in Lung cancer cells. Sci. Rep. 2016, 6, 37151. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Meng, Q.; Yang, M.; Liu, D.; Hou, X.; Tang, L.; Wang, X.; Lyu, Y.; Chen, X.; Liu, K.; et al. Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B. 2019, 9, 1113–1144. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, G.; Zhao, L.; Wang, S.; Kano, Y.; Yuan, D. Excretion of tectorigenin in rat urine orally administrated at different dosages by ultra-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry. Eur. J. Drug Metab. Pharmacokinet. 2015, 40, 255–266. [Google Scholar] [CrossRef]
- Qu, J.; Wu, Z.; Gao, J.; Wen, H.; Wang, T.; Yuan, D. Excretion of tectoridin metabolites in rat urine and bile orally administrated at different dosages and their inhibitory activity against aldose reductase. Fitoterapia 2014, 99, 99–108. [Google Scholar] [CrossRef]
- Hu, T.; Ge, X.; Wang, J.; Zhang, N.; Diao, X.; Hu, L.; Wang, X. Metabolite identification of iridin in rats by using UHPLC-MS/MS and pharmacokinetic study of its metabolite irigenin. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1181, 122914. [Google Scholar] [CrossRef]
- Wang, Q.; Hao, H.; Zhu, X.; Yu, G.; Lai, L.; Liu, Y.; Wang, Y.; Jiang, S.; Wang, G. Regioselective glucuronidation of tanshinone iia after quinone reduction: Identification of human UDP-glucuronosyltransferases, species differences, and interaction potential. Drug Metab. Dispos. 2010, 38, 1132–1140. [Google Scholar] [CrossRef]
- Shi, R.; Zhou, H.; Ma, B.; Ma, Y.; Wu, D.; Wang, X.; Luo, H.; Cheng, N. Pharmacokinetics and metabolism of jatrorrhizine, a gastric prokinetic drug candidate. Biopharm. Drug Dispos. 2012, 33, 135–145. [Google Scholar] [CrossRef]
- Sanchez-Dominguez, C.N.; Gallardo-Blanco, H.L.; Salinas-Santander, M.A.; Ortiz-Lopez, R. Uridine 5’-diphospho-glucronosyltrasferase: Its role in pharmacogenomics and human disease. Exp. Ther. Med. 2018, 16, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Mekonnen, T.F.; Panne, U.; Koch, M. Glucosylation and Glutathione Conjugation of Chlorpyrifos and Fluopyram Metabolites Using Electrochemistry/Mass Spectrometry. Molecules 2019, 24, 898. [Google Scholar] [CrossRef] [Green Version]
- Evans, W.E.; Relling, M.V. Pharmacogenomics: Translating functional genomics into rational therapeutics. Science 1999, 286, 487–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hong, X.; Yao, Z.; Dai, Y.; Zhao, G.; Qin, Z.; Wu, B.; Gonzalez, F.J.; Yao, X. Glucuronidation of icaritin by human liver microsomes, human intestine microsomes and expressed UDP-glucuronosyltransferase enzymes: Identification of UGT1A3, 1A9 and 2B7 as the main contributing enzymes. Xenobiotica 2018, 48, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Luo, F.; Wang, Y.; Jiang, H.; Wang, L.; Zhang, G.; Zhu, L.; Hu, M.; Wang, X.; Lu, L.; et al. Species-and gender-dependent differences in the glucuronidation of a flavonoid glucoside and its aglycone determined using expressed UGT enzymes and microsomes. Biopharm. Drug Dispos. 2015, 36, 622–635. [Google Scholar] [CrossRef]
- Wang, S.; Gong, T.; Lu, J.; Kano, Y.; Yuan, D. Simultaneous determination of tectorigenin and its metabolites in rat plasma by ultra performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013, 933, 50–58. [Google Scholar] [CrossRef]
- Rowland, A.; Miners, J.O.; Mackenzie, P.I. The UDP-glucuronosyltransferases: Their role in drug metabolism and detoxification. Int. J. Biochem. Cell Biol. 2013, 45, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Kiang, T.K.; Ensom, M.H.; Chang, T.K. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol. Ther. 2005, 106, 97–132. [Google Scholar] [CrossRef]
- Liu, J.; Xia, T.R. Identification of the metabolites produced following Iris tectorum Maxim oral administration and a network pharmacology-based analysis of their potential pharmacological properties. Xenobiotica 2021, 51, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Hitchings, R.; Kelly, L. Predicting and Understanding the Human Microbiome’s Impact on Pharmacology. Trends Pharmacol. Sci. 2019, 40, 495–505. [Google Scholar] [CrossRef]
- Luo, C.F.; Cai, B.; Hou, N.; Yuan, M.; Liu, S.M.; Ji, H.; Xiong, L.G.; Xiong, W.; Luo, J.D.; Chen, M.S. UDP-glucuronosyltransferase 1A1 is the principal enzyme responsible for puerarin metabolism in human liver microsomes. Arch. Toxicol. 2012, 86, 1681–1690. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, R.; Ma, B.; Ma, Y.; Wang, C.; Wu, D.; Wang, X.; Cheng, N. CYP450 1A2 and multiple UGT1A isoforms are responsible for jatrorrhizine metabolism in human liver microsomes. Biopharm. Drug Dispos. 2013, 34, 176–185. [Google Scholar] [CrossRef]
- Aprile, S.; Del Grosso, E.; Grosa, G. Identification of the human UDP-glucuronosyltransferases involved in the glucuronidation of combretastatin A-4. Drug Metab. Dispos. 2010, 38, 1141–1146. [Google Scholar] [CrossRef] [Green Version]
- Gradinaru, J.; Romand, S.; Geiser, L.; Carrupt, P.A.; Spaggiari, D.; Rudaz, S. Inhibition screening method of microsomal UGTs using the cocktail approach. Eur. J. Pharm. Sci. 2015, 71, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.Y.; Lee, H.; Lim, S.R.; Kim, J.H.; Lee, H.S. Effect of efavirenz on UDP-glucuronosyltransferase 1A1, 1A4, 1A6, and 1A9 activities in human liver microsomes. Molecules 2012, 17, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Huang, B.; Gu, T.; Tai, T.; Zhou, H.; Jia, Y.; Mi, Q.; Zhang, M.; Xie, H. Human UGT2B7 is the major isoform responsible for the glucuronidation of clopidogrel carboxylate. Biopharm. Drug Dispos. 2018, 39, 88–98. [Google Scholar] [CrossRef] [PubMed]
| Response Factors | Mean ± SE | Mean | |
|---|---|---|---|
| Atectorigenin/AM | Low | 0.91 ± 0.02 | 0.92 |
| High | 0.92 ± 0.02 | ||
| Airigenin/AM1 | Low | 0.87± 0.04 | 0.87 |
| High | 0.87 ± 0.06 | ||
| Airigenin/AM2 | Low | 0.88 ± 0.04 | 0.90 |
| High | 0.91 ± 0.06 | ||
| Compounds | Concentration | in Inactivated Metabolic System for 2 h | −80 °C for 7 Days | in Autosampler for 12 h | |||
|---|---|---|---|---|---|---|---|
| RSD (%) | RE (%) | RSD (%) | RE (%) | RSD (%) | RE (%) | ||
| M | Low | 2.13 | 0.38 | 3.02 | 2.90 | 5.27 | −2.13 |
| High | 2.08 | 0.59 | 1.97 | 4.71 | 1.08 | −5.18 | |
| M1 | Low | 3.21 | −3.43 | 2.86 | −2.01 | 3.41 | −2.40 |
| High | 1.09 | 2.10 | 6.23 | 1.84 | 2.15 | −3.06 | |
| M2 | Low | 4.32 | 1.89 | 3.04 | −2.85 | 5.57 | −2.98 |
| High | 1.92 | −2.07 | 1.95 | −3.49 | 3.29 | −5.07 | |
| Substrate | Enzyme | Km | Vmax |
|---|---|---|---|
| (μM) | (nmol/min/mg Protein) | ||
| Tectorigenin | UGT1A1 | 40.64 ± 5.56 | 9.01 ± 0.39 |
| UGT1A9 | 18.37 ± 1.16 | 3.49 ± 0.58 | |
| Irigenin | UGT1A1(M1) | 20.86 ± 2.04 | 5.22 ± 0.14 |
| UGT1A9(M1) | 73.88 ± 4.04 | 0.45 ± 0.09 | |
| UGT1A9(M2) | 25.04 ± 2.37 | 4.56 ± 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Xu, Z.; Gu, J. UGT1A1 and UGT1A9 Are Responsible for Phase II Metabolism of Tectorigenin and Irigenin In Vitro. Molecules 2022, 27, 4104. https://doi.org/10.3390/molecules27134104
Li J, Xu Z, Gu J. UGT1A1 and UGT1A9 Are Responsible for Phase II Metabolism of Tectorigenin and Irigenin In Vitro. Molecules. 2022; 27(13):4104. https://doi.org/10.3390/molecules27134104
Chicago/Turabian StyleLi, Ji, Zhangyao Xu, and Jifeng Gu. 2022. "UGT1A1 and UGT1A9 Are Responsible for Phase II Metabolism of Tectorigenin and Irigenin In Vitro" Molecules 27, no. 13: 4104. https://doi.org/10.3390/molecules27134104
APA StyleLi, J., Xu, Z., & Gu, J. (2022). UGT1A1 and UGT1A9 Are Responsible for Phase II Metabolism of Tectorigenin and Irigenin In Vitro. Molecules, 27(13), 4104. https://doi.org/10.3390/molecules27134104





