Current Research on HIV Drug Resistance
A topical collection in Pathogens (ISSN 2076-0817). This collection belongs to the section "Immunological Responses and Immune Defense Mechanisms".
Submission Status: Closed | Viewed by 43568Editor
Interests: HIV drug resistance; HIV genetics; HIV molecular epidemiology; metagenomics; next-generation sequencing
Topical Collection Information
Dear Colleagues,
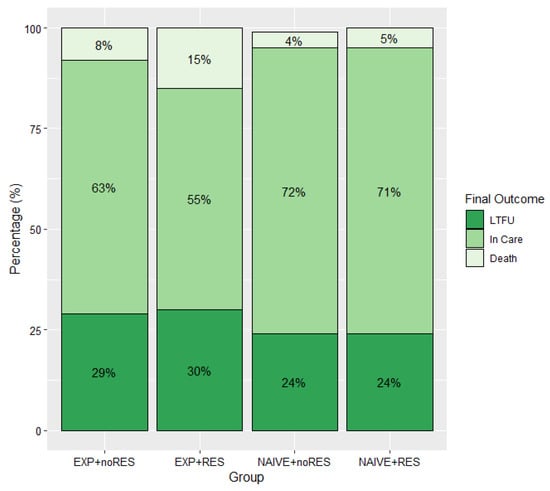
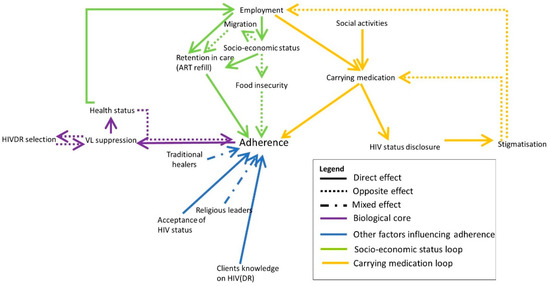
Nearly four decades have passed since the human immunodeficiency virus (HIV) was identified as the causal agent for acquired immunodeficiency syndrome (AIDS); however, HIV/AIDS remains one of the leading causes of death. This is especially the case in Sub-Saharan Africa, where the HIV/AIDS pandemic hit the hardest. While a preventative HIV vaccine remains intangible, we have witnessed remarkable progress in the development of antiretroviral therapy (ART) since 1987. With over 50 drugs currently available and the global scaling up of ART, HIV/AIDS has been successfully converted from a fatal disease into a manageable chronic infection when effective ART is readily accessible. Despite this, HIV drug resistance (HIVDR) has long been a major obstacle hindering the maximization of the clinical benefits from ART. The notable increase in HIVDR prevalence, accompanying the global growth in ART usage, renders HIVDR an even more urgent issue to deal with for the sustainable reduction of HIV/AIDS-associated morbidity and mortality.
In this context, Pathogens has launched a thematic issue devoted to the “Current Research on HIV Drug Resistance”, aiming to publish the latest findings in this important research field. All submissions pertaining to HIVDR are welcome! The manuscript could be presented as an original research article, review, commentary or viewpoint piece, whichever is appropriate. Topics of particular interest include but are not limited to:
- HIVDR surveillance and epidemiology, and their applications in a public health context;
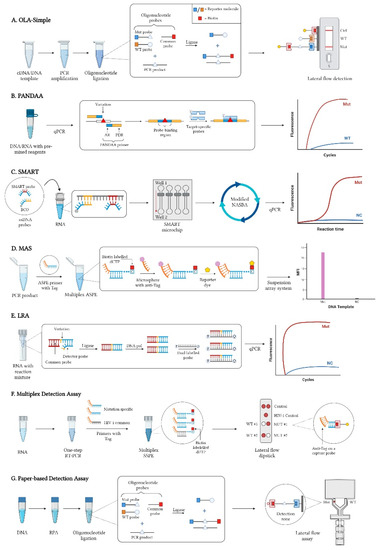
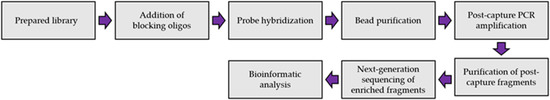
- Novel HIVDR testing technologies with improved sensitivity, accuracy and practicability;
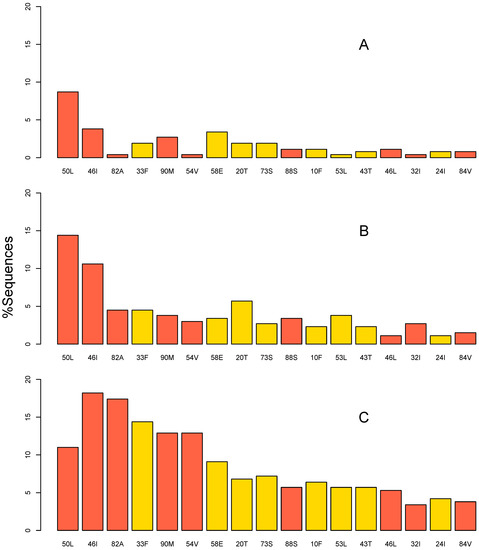
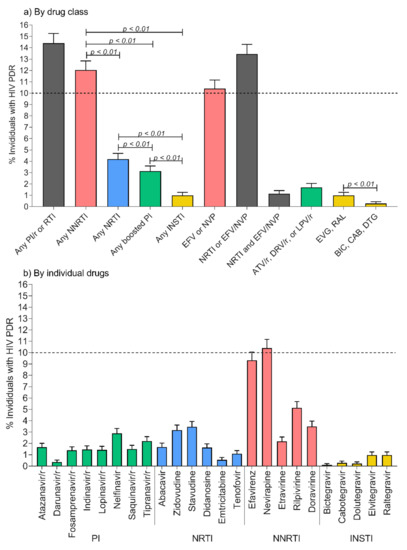
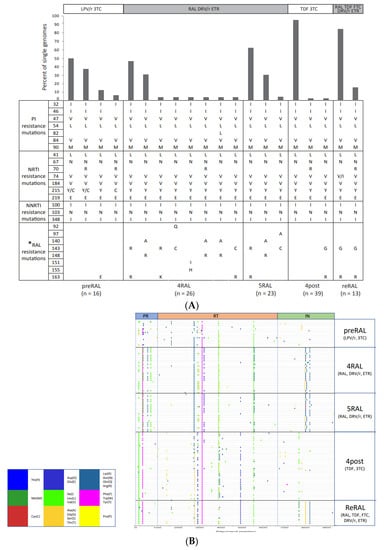
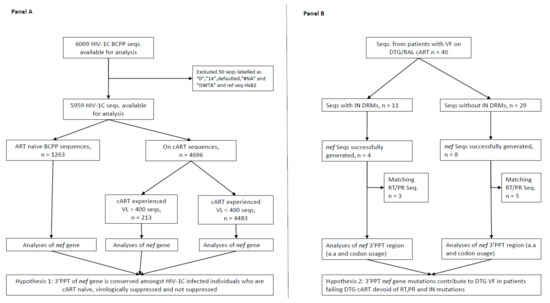
- HIVDR against integrase inhibitors and newly approved ART drugs;
- In-depth research on HIVDR mechanisms;
- The clinical management of HIVDR.
Prof. Dr. Hezhao Ji
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 250 words) can be sent to the Editorial Office for assessment.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Pathogens is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2200 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- HIV
- drug resistance
- research
- laboratory testing
- clinical management
- surveillance