Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications
Abstract
:Simple Summary
Abstract
1. Introduction
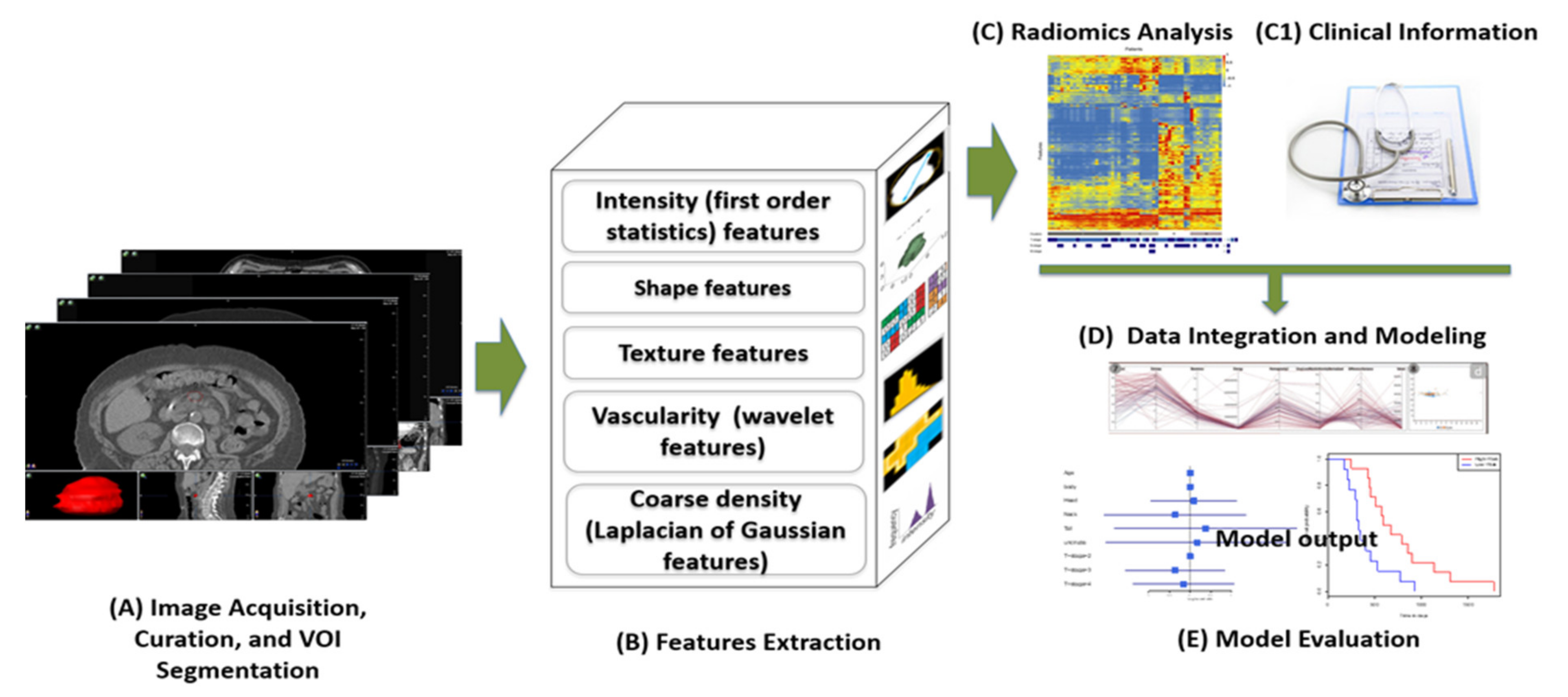
2. Technical Overview: Quantitative Imaging and Two Analytical Approaches
2.1. Technical Basis: Radiomics
2.2. Technical Basis: Deep Learning
3. Clinical Applications
3.1. Pre-Cancerous Pancreatic Lesion Diagnosis
3.2. Pancreatic Cancer Detection and Diagnosis
3.3. Pancreatic Cancer Prognosis
3.4. Treatment Stratification, Delta-Radiomics, and Radiogenomics
4. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- American Cancer Society: Cancer Facts & Statistics. Available online: https://cancerstatisticscenter.cancer.org/?_ga=2.62302948.97622418.1643164702-1977482543.1643164701#!/cancer-site/Pancreas (accessed on 26 January 2022).
- Chiaro, M.D. Early Detection and Prevention of Pancreatic Cancer: Is It Really Possible Today? World J. Gastroenterol. 2014, 20, 12118. [Google Scholar] [PubMed]
- Peluso, H.; Jones, W.B.; Parikh, A.A.; Abougergi, M.S. Treatment Outcomes, 30-Day Readmission and Healthcare Resource Utilization after Pancreatoduodenectomy for Pancreatic Malignancies. J. Hepato-Biliary-Pancreat. Sci. 2019, 26, 187–194. [Google Scholar]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The Facts and the Challenges of Image Analysis. Eur. Radiol. Exp. 2018, 2, 1–8. [Google Scholar]
- Avanzo, M.; Stancanello, J.; Pirrone, G.; Sartor, G. Radiomics and deep learning in lung cancer. Strahlenther. Onkol. 2020, 196, 879–887. [Google Scholar] [CrossRef]
- Thawani, R.; McLane, M.; Beig, N.; Ghose, S.; Prasanna, P.; Velcheti, V.; Madabhushi, A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018, 115, 34–41. [Google Scholar] [CrossRef]
- Liang, Y.; Schott, D.; Zhang, Y.; Wang, Z.; Nasief, H.; Paulson, E.; Hall, W.; Knechtges, P.; Erickson, B.; Li, X.A. Auto-segmentation of pancreatic tumor in multi-parametric MRI using deep convolutional neural networks. Radiother. Oncol. 2020, 145, 193–200. [Google Scholar] [CrossRef]
- Lim, C.H.; Cho, Y.S.; Choi, J.Y.; Lee, K.-H.; Lee, J.K.; Min, J.H.; Hyun, S.H. Imaging Phenotype Using 18F-Fluorodeoxyglucose Positron Emission Tomography–Based Radiomics and Genetic Alterations of Pancreatic Ductal Adeno-Carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2113–2122. [Google Scholar]
- Iwatate, Y.; Hoshino, I.; Yokota, H.; Ishige, F.; Itami, M.; Mori, Y.; Chiba, S.; Arimitsu, H.; Yanagibashi, H.; Nagase, H.; et al. Radiogenomics for predicting p53 status, PD-L1 expression, and prognosis with machine learning in pancreatic cancer. Br. J. Cancer 2020, 123, 1253–1261. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and Deep Learning Methods for Radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar]
- Kriegeskorte, N.; Golan, T. Neural network models and deep learning. Curr. Biol. 2019, 29, R231–R236. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. NIPS 2012, 60, 84–90. [Google Scholar]
- Pan, S.J.; Yang, Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar]
- Tas, F.; Sen, F.; Keskin, S.; Kilic, L.; Yildiz, I. Prognostic Factors in Metastatic Pancreatic Cancer: Older Patients Are Associated with Reduced Overall Survival. Mol. Clin. Oncol. 2013, 1, 788–792. [Google Scholar] [PubMed] [Green Version]
- Pancreatic Cancer—Statistics. Available online: https://www.cancer.net/cancer-types/pancreatic-cancer/statistics (accessed on 21 January 2022).
- Goggins, M.; Overbeek, K.A.; Brand, R.; Syngal, S.; Del Chiaro, M.; Bartsch, D.K.; Bassi, C.; Carrato, A.; Farrell, J.; Fishman, E.K.; et al. Management of Patients with In-creased Risk for Familial Pancreatic Cancer: Updated Recommendations from the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut 2019, 69, 7–17. [Google Scholar]
- Hanania, A.N.; Bantis, L.E.; Feng, Z.; Wang, H.; Tamm, E.P.; Katz, M.H.; Maitra, A.; Koay, E.J. Quantitative imaging to evaluate malignant potential of IPMNs. Oncotarget 2016, 7, 85776–85784. [Google Scholar] [CrossRef] [Green Version]
- Tobaly, D.; Santinha, J.; Sartoris, R.; Dioguardi Burgio, M.; Matos, C.; Cros, J.; Couvelard, A.; Rebours, V.; Sauvanet, A.; Ro-not, M.; et al. CT-Based Radiomics Analysis to Predict Malignancy in Patients with Intraductal Papillary Mucinous Neoplasm (IPMN) of the Pancreas. Cancers 2020, 12, 3089. [Google Scholar]
- Permuth, J.B.; Choi, J.; Balarunathan, Y.; Kim, J.; Chen, D.-T.; Chen, L.; Orcutt, S.; Doepker, M.P.; Gage, K.; Zhang, G.; et al. Combining radiomic features with a miRNA classifier may improve prediction of malignant pathology for pancreatic intraductal papillary mucinous neoplasms. Oncotarget 2016, 7, 85785–85797. [Google Scholar] [CrossRef] [Green Version]
- Wei, R.; Lin, K.; Yan, W.; Guo, Y.; Wang, Y.; Li, J.; Zhu, J. Computer-Aided Diagnosis of Pancreas Serous Cystic Neoplasms: A Radiomics Method on Preoperative MDCT Images. Technol. Cancer Res. Treat. 2019, 18, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Ma, S.; Guo, X.; Zhang, X.; Wang, X. Preoperative differentiation of pancreatic mucinous cystic neoplasm from macrocystic serous cystic adenoma using radiomics: Preliminary findings and comparison with radiological model. Eur. J. Radiol. 2020, 122, 108747. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, J.; Midya, A.; Gazit, L.; Attiyeh, M.; Langdon-Embry, L.; Allen, P.J.; Do, R.K.; Simpson, A.L. CT Radiomics to Predict High-Risk Intraductal Papillary Mucinous Neoplasms of the Pancreas. Med Phys. 2018, 45, 5019–5029. [Google Scholar]
- Harrington, K.A.; Williams, T.L.; Lawrence, S.A.; Chakraborty, J.; Al Efishat, M.A.; Attiyeh, M.A.; Askan, G.; Chou, Y.; Pulvirenti, A.; McIntyre, C.A.; et al. Multimodal radiomics and cyst fluid inflammatory markers model to predict preoperative risk in intraductal papillary mucinous neoplasms. J. Med. Imaging 2020, 7, 031507. [Google Scholar] [CrossRef]
- Shen, X.; Yang, F.; Yang, P.; Yang, M.; Xu, L.; Zhuo, J.; Wang, J.; Lu, D.; Liu, Z.; Zheng, S.-S.; et al. A Contrast-Enhanced Computed Tomography Based Radiomics Approach for Preoperative Differentiation of Pancreatic Cystic Neo-Plasm Subtypes: A Feasibility Study. Front. Oncol. 2020, 10, 248. [Google Scholar] [PubMed] [Green Version]
- Chen, S.; Ren, S.; Guo, K.; Daniels, M.J.; Wang, Z.; Chen, R. Preoperative differentiation of serous cystic neoplasms from mucin-producing pancreatic cystic neoplasms using a CT-based radiomics nomogram. Abdom. Radiol. 2021, 46, 2637–2646. [Google Scholar] [CrossRef]
- Cui, S.; Tang, T.; Su, Q.; Wang, Y.; Shu, Z.; Yang, W.; Gong, X. Radiomic nomogram based on MRI to predict grade of branching type intraductal papillary mucinous neoplasms of the pancreas: A multicenter study. Cancer Imaging 2021, 21, 1–13. [Google Scholar] [CrossRef]
- Xie, T.; Wang, X.; Zhang, Z.; Zhou, Z. CT-Based Radiomics Analysis for Preoperative Diagnosis of Pancreatic Mucinous Cyst-ic Neoplasm and Atypical Serous Cystadenomas. Front. Oncol. 2021, 11, 621520. [Google Scholar] [CrossRef] [PubMed]
- Polk, S.L.; Choi, J.W.; Mcgettigan, M.J.; Rose, T.; Ahmed, A.; Kim, J.; Jiang, K.; Balagurunathan, Y.; Qi, J.; Farah, P.T.; et al. Multiphase computed tomography radiomics of pancreatic intraductal papillary mucinous neoplasms to predict malignancy. World J. Gastroenterol. 2020, 26, 3458–3471. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Shi, H.; Lu, M.; Wang, C.; Duan, S.; Xu, Q.; Shi, H. Radiomics Analysis for Predicting Malignant Potential of In-traductal Papillary Mucinous Neoplasms of the Pancreas: Comparison of CT and MRI. Acad. Radiol. 2021, 29, 367–375. [Google Scholar] [CrossRef] [PubMed]
- D’Onofrio, M.; Tedesco, G.; Cardobi, N.; De Robertis, R.; Sarno, A.; Capelli, P.; Martini, P.T.; Giannotti, G.; Beleù, A.; Marchegiani, G.; et al. Magnetic resonance (MR) for mural nodule detection studying Intraductal papillary mucinous neoplasms (IPMN) of pancreas: Imaging-pathologic correlation. Pancreatology 2020, 21, 180–187. [Google Scholar] [CrossRef]
- Sahani, D.V.; Sainani, N.I.; Blake, M.A.; Crippa, S.; Mino-Kenudson, M.; del-Castillo, C.F. Prospective Evaluation of Read-er Performance on MDCT in Characterization of Cystic Pancreatic Lesions and Prediction of Cyst Biologic Aggressiveness. Am. J. Roentgenol. 2011, 197, W53–W61. [Google Scholar] [CrossRef]
- Huang, W.-P.; Liu, S.-Y.; Han, Y.-J.; Li, L.-M.; Liang, P.; Gao, J.-B. Development of CT-Based Imaging Signature for Preoperative Prediction of Invasive Behavior in Pancreatic Solid Pseudopapillary Neoplasm. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Song, C.; Wang, M.; Luo, Y.; Chen, J.; Peng, Z.; Wang, Y.; Zhang, H.; Li, Z.-P.; Shen, J.; Huang, B.; et al. Predicting the Recurrence Risk of Pancreatic Neuroendocrine Neoplasms after Radical Resection Using Deep Learning Radiomics with Pre-operative Computed Tomography Images. Ann. Transl. Med. 2021, 9, 833. [Google Scholar] [PubMed]
- Watson, M.D.; Lyman, W.B.; Passeri, M.J.; Murphy, K.J.; Sarantou, J.P.; Iannitti, D.A.; Martinie, J.B.; Vrochides, D.; Baker, E.H. Use of Artificial Intelligence Deep Learning to Determine the Malignant Potential of Pancreatic Cystic Neo-Plasms with Preoperative Computed Tomography Imaging. Am. Surg. 2020, 87, 602–607. [Google Scholar] [PubMed]
- Awe, A.M.; Rendell, V.R.; Lubner, M.G.; Winslow, E.R. Texture Analysis. Pancreas 2020, 49, 301–312. [Google Scholar] [PubMed]
- Attiyeh, M.A.; Chakraborty, J.; McIntyre, C.A.; Kappagantula, R.; Chou, Y.; Askan, G.; Seier, K.; Gonen, M.; Basturk, O.; Balachandran, V.P.; et al. CT Radiomics Associations with Genotype and Stromal Content in Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. 2019, 44, 3148–3157. [Google Scholar]
- Yang, J.; Guo, X.; Ou, X.; Zhang, W.; Ma, X. Discrimination of Pancreatic Serous Cystadenomas from Mucinous Cystadeno-mas With CT Textural Features: Based on Machine Learning. Front. Oncol. 2019, 9, 494. [Google Scholar] [CrossRef] [Green Version]
- Kurita, Y.; Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Koda, H.; Tajika, M.; Shimizu, Y.; et al. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci. Rep. 2019, 9, 6893. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Liu, Z.-X.; Zhang, J.-J.; Wu, F.-T.; Xu, C.-F.; Shen, Z.; Yu, C.-H.; Li, Y.-M. Construction of a convolutional neural network classifier developed by computed tomography images for pancreatic cancer diagnosis. World J. Gastroenterol. 2020, 26, 5156–5168. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, X.; Chen, J.; Song, C.; Shen, J.; Xiao, H.; Chen, M.; Li, Z.-P.; Huang, B.; Feng, S.-T. Preoperative Prediction of Pancreatic Neuroendocrine Neoplasms Grading Based on Enhanced Computed Tomography Imaging: Validation of Deep Learning with a Convolutional Neural Network. Neuroendocrinology 2019, 110, 338–350. [Google Scholar] [CrossRef]
- Dmitriev, K.; Kaufman, A.E.; Javed, A.A.; Hruban, R.H.; Fishman, E.K.; Lennon, A.M.; Saltz, J.H. Classification of Pancreatic Cysts in Computed Tomography Images Using a Random Forest and Convolutional Neural Network Ensemble. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2017; pp. 150–158. [Google Scholar]
- Corral, J.E.; Hussein, S.; Kandel, P.; Bolan, C.W.; Bagci, U.; Wallace, M.B. Deep Learning to Classify Intraductal Papillary Mucinous Neoplasms Using Magnetic Resonance Imaging. Pancreas 2019, 48, 805–810. [Google Scholar] [CrossRef]
- Kuwahara, T.; Hara, K.; Mizuno, N.; Okuno, N.; Matsumoto, S.; Obata, M.; Kurita, Y.; Koda, H.; Toriyama, K.; Onishi, S.; et al. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin. Transl. Gastroenterol. 2019, 10, e00045. [Google Scholar] [CrossRef]
- Abel, L.; Wasserthal, J.; Weikert, T.; Sauter, A.W.; Nesic, I.; Obradovic, M.; Yang, S.; Manneck, S.; Glessgen, C.; Ospel, J.M.; et al. Automated Detection of Pancreatic Cystic Lesions on CT Using Deep Learning. Diagnostics 2021, 11, 901. [Google Scholar] [CrossRef]
- Nguon, L.S.; Seo, K.; Lim, J.-H.; Song, T.-J.; Cho, S.-H.; Park, J.-S.; Park, S. Deep Learning-Based Differentiation between Mucinous Cystic Neoplasm and Serous Cystic Neoplasm in the Pancreas Using Endoscopic Ultrasonography. Diagnostics 2021, 11, 1052. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Chen, Y.; Sa, G.; Li, K.; Hu, H.; Zhou, J.; Guan, Q.; Chen, F. CT classification model of pancreatic serous cystic neoplasms and mucinous cystic neoplasms based on a deep neural network. Abdom. Radiol. 2022, 47, 232–241. [Google Scholar] [CrossRef]
- Chu, L.; Goggins, M.G.; Fishman, E. Diagnosis and Detection of Pancreatic Cancer. Cancer J. 2017, 23, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, G.; Mori, M.; Panzeri, M.M.; Barbera, M.; Palumbo, D.; Sini, C.; Muffatti, F.; Andreasi, V.; Steidler, S.; Doglioni, C.; et al. CT-derived radiomic features to discriminate histologic characteristics of pancreatic neuroendocrine tumors. Radiol. Med. 2021, 126, 745–760. [Google Scholar] [CrossRef]
- Bevilacqua, A.; Calabrò, D.; Malavasi, S.; Ricci, C.; Casadei, R.; Campana, D.; Baiocco, S.; Fanti, S.; Ambrosini, V. A [68Ga]Ga-DOTANOC PET/CT Radiomic Model for Non-Invasive Prediction of Tumour Grade in Pancreatic Neuroendocrine Tumours. Diagnostics 2021, 11, 870. [Google Scholar] [CrossRef]
- Bian, Y.; Li, J.; Cao, K.; Fang, X.; Jiang, H.; Ma, C.; Jin, G.; Lu, J.; Wang, L. Magnetic resonance imaging radiomic analysis can preoperatively predict G1 and G2/3 grades in patients with NF-pNETs. Abdom. Radiol. 2021, 46, 667–680. [Google Scholar] [CrossRef]
- Bian, Y.; Zhao, Z.; Jiang, H.; Fang, H.; Li, J.; Cao, K.; Ma, C.; Guo, S.; Wang, L.; Jin, G.; et al. Noncontrast Radiomics Approach for Predicting Grades of Nonfunctional Pancreatic Neuroendocrine Tumors. J. Mag. Res. Imaging 2020, 52, 1124–1136. [Google Scholar] [CrossRef]
- Canellas, R.; Burk, K.S.; Parakh, A.; Sahani, D.V. Prediction of Pancreatic Neuroendocrine Tumor Grade Based on CT Features and Texture Analysis. Am. J. Roentgenol. 2018, 210, 341–346. [Google Scholar]
- Chang, N.; Cui, L.; Luo, Y.; Chang, Z.; Yu, B.; Liu, Z. Development and Multicenter Validation of a CT-Based Radiomics Sig-nature for Discriminating Histological Grades of Pancreatic Ductal Adenocarcinoma. Quant. Imaging Med. Surg. 2020, 10, 692–702. [Google Scholar]
- Chen, P.-T.; Chang, D.; Yen, H.; Liu, K.-L.; Huang, S.-Y.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Radiomic Features at CT Can Distinguish Pancreatic Cancer from Noncancerous Pancreas. Radiol. Imaging Cancer 2021, 3, e210010. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.C.; Park, S.; Kawamoto, S.; Fouladi, D.F.; Shayesteh, S.; Zinreich, E.S.; Graves, J.S.; Horton, K.M.; Hruban, R.H.; Yuille, A.L.; et al. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma from Normal Pancreatic Tissue. Am. J. Roentgenol. 2019, 213, 349–357. [Google Scholar] [CrossRef]
- Deng, Y.; Ming, B.; Zhou, T.; Wu, J.; Chen, Y.; Liu, P.; Zhang, J.; Zhang, S.; Chen, T.; Zhang, X.-M. Radiomics Model Based on MR Images to Discriminate Pancreatic Ductal Adenocarcinoma and Mass-Forming Chronic Pancreatitis Lesions. Front. Oncol. 2021, 11, 811. [Google Scholar]
- Gu, H.; Liang, H.; Zhong, J.; Wei, Y.; Ma, Y. How Does the Pancreatic Solid Pseudopapillary Neoplasm Confuse Us: Analyzing from the Point View of MRI-Based Radiomics? Magn. Reson. Imaging 2022, 85, 38–43. [Google Scholar] [PubMed]
- Li, J.; Lu, J.; Liang, P.; Li, A.; Hu, Y.; Shen, Y.; Hu, D.; Li, Z. Differentiation of atypical pancreatic neuroendocrine tumors from pancreatic ductal adenocarcinomas: Using whole-tumor CT texture analysis as quantitative biomarkers. Cancer Med. 2018, 7, 4924–4931. [Google Scholar] [CrossRef]
- Linning, E.; Xu, Y.; Wu, Z.; Li, L.; Zhang, N.; Yang, H. Differentiation of Focal-Type Autoimmune Pancreatitis from Pancreatic Ductal Adenocarcinoma Using Radiomics Based on Multiphasic Computed. J. Comput. Assist. Tomogr. 2020, 44, 511–518. [Google Scholar] [CrossRef]
- Liu, Z.; Li, M.; Zuo, C.; Yang, Z.; Yang, X.; Ren, S.; Peng, Y.; Sun, G.; Shen, J.; Cheng, C.; et al. Radiomics model of dual-time 2-[18F]FDG PET/CT imaging to distinguish between pancreatic ductal adenocarcinoma and autoimmune pancreatitis. Eur. Radiol. 2021, 31, 6983–6991. [Google Scholar] [CrossRef]
- Liu, C.; Bian, Y.; Meng, Y.; Liu, F.; Cao, K.; Zhang, H.; Fang, X.; Li, J.; Yu, J.; Feng, X.; et al. Preoperative Prediction of G1 and G2/3 Grades in Patients with Nonfunctional Pancreatic Neuroendocrine Tumors Using Multimodality Imaging. Acad. Radiol. 2021, 29, 49–60. [Google Scholar] [CrossRef]
- Park, S.; Chua, L.C.; Hrubanbc, R.H.; Vogelsteincde, B.; Kinzlercd, K.W.; Yuillefg, A.L.; Fouladia, D.F.; Shayesteha, S.; Ghandilia, S.; Wolfgangh, C.L.; et al. Differentiating autoimmune pancreatitis from pancreatic ductal adenocarcinoma with CT radiomics features. Diagn. Interv. Imaging 2020, 101, 555–564. [Google Scholar] [CrossRef]
- Reinert, C.P.; Baumgartner, K.; Hepp, T.; Bitzer, M.; Horger, M. Complementary role of computed tomography texture analysis for differentiation of pancreatic ductal adenocarcinoma from pancreatic neuroendocrine tumors in the portal-venous enhancement phase. Abdom. Radiol. 2020, 45, 750–758. [Google Scholar] [CrossRef]
- Ren, S.; Zhao, R.; Cui, W.; Qiu, W.; Guo, K.; Cao, Y.; Duan, S.; Wang, Z.; Chen, R. Computed Tomography-Based Radiomics Signature for the Preoperative Differentiation of Pancreatic Adenosquamous Carcinoma from Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2020, 10, 1618. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Zhang, Q.-W.; Duan, S.-F.; Bian, Y.; Hao, Q.; Xing, P.-Y.; Wang, T.-G.; Chen, L.-G.; Ma, C.; Lu, J.-P. MRI-Based Radiomics Approach for Differentiation of Hypovascular Non-Functional Pancreatic Neuroendocrine Tumors and Solid Pseudopapillary Neoplasms of the Pancreas. BMC Med. Imaging 2021, 21, 36. [Google Scholar] [CrossRef]
- Xing, H.; Hao, Z.; Zhu, W.; Sun, D.; Ding, J.; Zhang, H.; Liu, Y.; Huo, L. Preoperative Prediction of Pathological Grade in Pancreatic Ductal Adenocarcinoma Based on 18F-FDG PET/CT Radiomics. EJNMMI Res. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Y.H.; Liu, X.; Xu, H.; Chen, C.; Zhou, X.; Liu, Y.; Ma, X. Application of Radiomics Analysis Based on CT Combined with Machine Learning in Diagnostic of Pancreatic Neuroendocrine Tumors Patient’s Pathological Grades. Front. Oncol. 2021, 10, 3227. [Google Scholar]
- Zhao, Z.; Bian, Y.; Jiang, H.; Fang, X.; Li, J.; Cao, K.; Ma, C.; Wang, L.; Zheng, J.; Yue, X.; et al. CT-Radiomic Approach to Predict G1/2 Nonfunctional Pancreatic Neuroendocrine Tumor. Acad. Radiol. 2020, 27, e272–e281. [Google Scholar]
- Chu, L.C.; Park, S.; Kawamoto, S.; Wang, Y.; Zhou, Y.; Shen, W.; Zhu, Z.; Xia, Y.; Xie, L.; Liu, F.; et al. Application of Deep Learning to Pancreatic Cancer Detection: Lessons Learned from Our Initial Experience. J. Am. Coll. Radiol. 2019, 16, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-L.; Wu, T.; Chen, P.-T.; Tsai, Y.M.; Roth, H.; Wu, M.-S.; Liao, W.-C.; Wang, W. Deep Learning to Distinguish Pancreatic Cancer Tissue from Non-Cancerous Pancreatic Tissue: A Retrospective Study with Cross-Racial External Validation. Lancet Digit. Health 2020, 2, e303–e313. [Google Scholar]
- Ozkan, M.; Cakiroglu, M.; Kocaman, O.; Kurt, M.; Yilmaz, B.; Can, G.; Korkmaz, U.; Dandil, E.; Eksi, Z. Age-Based Computer-Aided Diagnosis Approach for Pancreatic Cancer on Endoscopic Ultrasound Images. Endosc. Ultrasound 2016, 5, 101. [Google Scholar] [PubMed] [Green Version]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Gheonea, D.I.; Gorunescu, M.; Ciurea, T.; Popescu, G.L.; Iordache, A.; Hassan, H.; Iordache, S. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest. Endosc. 2008, 68, 1086–1094. [Google Scholar] [CrossRef]
- Săftoiu, A.; Vilmann, P.; Gorunescu, F.; Janssen, J.; Hocke, M.; Larsen, M.; Iglesias–Garcia, J.; Arcidiacono, P.; Will, U.; Giovannini, M.; et al. Efficacy of an Artificial Neural Network–Based Approach to Endoscopic Ultrasound Elastography in Diagnosis of Focal Pancreatic Masses. Clin. Gastroenterol. Hepatol. 2012, 10, 1. [Google Scholar]
- Si, K.; Xue, Y.; Yu, X.; Zhu, X.; Li, Q.; Gong, W.; Liang, T.; Duan, S. Fully End-to-End Deep-Learning-Based Diagnosis of Pancreatic Tumors. Theranostics 2021, 11, 1982–1990. [Google Scholar] [PubMed]
- Tonozuka, R.; Itoi, T.; Nagata, N.; Kojima, H.; Sofuni, A.; Tsuchiya, T.; Ishii, K.; Tanaka, R.; Nagakawa, Y.; Mukai, S. Deep Learning Analysis for the Detection of Pancreatic Cancer on Endosonographic Images: A Pilot Study. J. Hepato-Biliary-Pancreat. Sci. 2020, 28, 95–104. [Google Scholar]
- Zhang, Z.; Li, S.; Wang, Z.; Lu, Y. A Novel and Efficient Tumor Detection Framework for Pancreatic Cancer via CT Images. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 30 October 2020. [Google Scholar]
- Zhang, Z.; Li, S.; Wang, Z.; Lu, Y. A Novel and Efficient Tumor Detection Framework for Pancreatic Cancer via CT Images. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2020, 1160–1164. [Google Scholar] [CrossRef]
- Zhu, M.; Xu, C.; Yu, J.; Wu, Y.; Li, C.; Zhang, M.; Jin, Z.; Li, Z. Differentiation of Pancreatic Cancer and Chronic Pancreatitis Using Computer-Aided Diagnosis of Endoscopic Ultrasound (EUS) Images: A Diagnostic Test. PLoS ONE 2013, 8, e63820. [Google Scholar] [CrossRef] [Green Version]
- Ziegelmayer, S.; Kaissis, G.; Harder, F.; Jungmann, F.; Müller, T.; Makowski, M.; Braren, R. Deep Convolutional Neural Network-Assisted Feature Extraction for Diagnostic Discrimination and Feature Visualization in Pancreatic Ductal Adenocarcinoma (PDAC) versus Autoimmune Pancreatitis (AIP). J. Clin. Med. 2020, 9, 4013. [Google Scholar]
- Cheng, S.-H.; Cheng, Y.-J.; Jin, Z.-Y.; Xue, H.-D. Unresectable pancreatic ductal adenocarcinoma: Role of CT quantitative imaging biomarkers for predicting outcomes of patients treated with chemotherapy. Eur. J. Radiol. 2019, 113, 188–197. [Google Scholar] [CrossRef]
- Parr, E.; Du, Q.; Zhang, C.; Lin, C.; Kamal, A.; McAlister, J.; Liang, X.; Bavitz, K.; Rux, G.; Hollingsworth, M.; et al. Radiomics-Based Outcome Prediction for Pancreatic Cancer Following Stereotactic Body Radiotherapy. Cancers 2020, 12, 1051. [Google Scholar] [CrossRef]
- Li, K.; Yao, Q.; Xiao, J.; Li, M.; Yang, J.; Hou, W.; Du, M.; Chen, K.; Qu, Y.; Li, L.; et al. Contrast-Enhanced CT Radiomics for Predicting Lymph Node Metastasis in Pancreatic Ductal Adenocarcinoma: A Pilot Study. Cancer Imaging 2020, 20, 1–10. [Google Scholar]
- Cusumano, D.; Boldrini, L.; Yadav, P.; Casà, C.; Lee, S.; Romano, A.; Piras, A.; Chiloiro, G.; Placidi, L.; Catucci, F.; et al. Delta Radiomics Analysis for Local Control Prediction in Pancreatic Cancer Patients Treated Using Magnetic Resonance Guided Radiotherapy. Diagnostics 2021, 11, 72. [Google Scholar] [CrossRef]
- Cen, C.; Liu, L.; Li, X.; Wu, A.; Liu, H.; Wang, X.; Wu, H.; Wang, C.; Han, P.; Wang, S. Pancreatic Ductal Adenocarcinoma at CT: A Combined Nomogram Model to Preoperatively Predict Cancer Stage and Survival Outcome. Front. Oncol. 2021, 11, 1980. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, Y.J.; Kim, K.G.; Park, J.S. Preoperative CT texture features predict prognosis after curative resection in pancreatic cancer. Sci. Rep. 2019, 9, 17389. [Google Scholar] [CrossRef] [PubMed]
- Khalvati, F.; Zhang, Y.; Baig, S.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A. Prognostic Value of CT Radiomic Features in Resectable Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2019, 9, 5449. [Google Scholar] [CrossRef] [PubMed]
- Toyama, Y.; Hotta, M.; Motoi, F.; Takanami, K.; Minamimoto, R.; Takase, K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Sci. Rep. 2020, 10, 17024. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Wang, X.; Li, M.; Tong, T.; Yu, X.; Zhou, Z. Pancreatic Ductal Adenocarcinoma: A Radiomics Nomogram Outperforms Clinical Model and TNM Staging for Survival Estimation after Curative Resection. Eur. Radiol. 2020, 30, 2513–2524. [Google Scholar] [PubMed]
- Hang, J.; Xu, K.; Yin, R.; Shao, Y.; Liu, M.; Shi, H.; Wang, X.; Wu, L. Role of CT texture features for predicting outcome of pancreatic cancer patients with liver metastases. J. Cancer 2021, 12, 2351–2358. [Google Scholar] [CrossRef]
- Mori, M.; Passoni, P.; Incerti, E.; Bettinardi, V.; Broggi, S.; Reni, M.; Whybra, P.; Spezi, E.; Vanoli, E.G.; Gianolli, L.; et al. Training and validation of a robust PET radiomic-based index to predict distant-relapse-free-survival after radio-chemotherapy for locally advanced pancreatic cancer. Radiother. Oncol. 2020, 153, 258–264. [Google Scholar] [CrossRef]
- Salinas-Miranda, E.; Khalvati, F.; Namdar, K.; Deniffel, D.; Dong, X.; Abbas, E.; Wilson, J.M.; O’Kane, G.M.; Knox, J.; Gallinger, S.; et al. Validation of Prognostic Radiomic Features from Resectable Pancreatic Ductal Adenocarcinoma in Patients with Advanced Disease Undergoing Chemotherapy. Can. Assoc. Radiol. J. 2020, 72, 605–613. [Google Scholar] [CrossRef]
- Li, K.; Xiao, J.; Yang, J.; Li, M.; Xiong, X.; Nian, Y.; Qiao, L.; Wang, H.; Eresen, A.; Zhang, Z.; et al. Association of radiomic imaging features and gene expression profile as prognostic factors in pancreatic ductal adenocarcinoma. Am. J. Transl. Res. 2019, 11, 4491–4499. [Google Scholar]
- D’Onofrio, M.; De Robertis, R.; Aluffi, G.; Cadore, C.; Beleù, A.; Cardobi, N.; Malleo, G.; Manfrin, E.; Bassi, C. CT Simplified Radiomic Approach to Assess the Metastatic Ductal Adenocarcinoma of the Pancreas. Cancers 2021, 13, 1843. [Google Scholar] [CrossRef]
- Mapelli, P.; Partelli, S.; Salgarello, M.; Doraku, J.; Muffatti, F.; Schiavo Lena, M.; Pasetto, S.; Bezzi, C.; Bettinardi, V.; Andreasi, V.; et al. Dual Tracer 68ga-DOTATOC and 18F-FDG Pet Improve Pre-operative Evaluation of Aggressiveness in Resectable Pancreatic Neuroendocrine Neoplasms. Diagnostics 2021, 11, 192. [Google Scholar]
- Kaissis, G.; Ziegelmayer, S.; Lohöfer, F.; Algül, H.; Eiber, M.; Weichert, W.; Schmid, R.; Friess, H.; Rummeny, E.; Ankerst, D.; et al. A Machine Learning Model for the Prediction of Survival and Tumor Subtype in Pancreatic Ductal Ad-enocarcinoma from Preoperative Diffusion-Weighted Imaging. Eur. Radiol. Exp. 2019, 3, 1–9. [Google Scholar]
- Yao, J.; Shi, Y.; Lu, L.; Xiao, J.; Zhang, L. DeepPrognosis: Preoperative Prediction of Pancreatic Cancer Survival and Surgical Margin via Contrast-Enhanced CT Imaging. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020. [Google Scholar] [CrossRef]
- Zhang, Y.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A.; Khalvati, F. Prognostic Value of Transfer Learn-ing Based Features in Resectable Pancreatic Ductal Adenocarcinoma. Front. Artif. Intell. 2020, 3, 77. [Google Scholar]
- Zhang, Y.; Lobo-Mueller, E.M.; Karanicolas, P.; Gallinger, S.; Haider, M.A.; Khalvati, F. Improving prognostic performance in resectable pancreatic ductal adenocarcinoma using radiomics and deep learning features fusion in CT images. Sci. Rep. 2021, 11, 1378. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Wang, X. Deep learning for World Health Organization grades of pancreatic neuroendocrine tumors on contrast-enhanced magnetic resonance images: A preliminary study. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1981–1991. [Google Scholar] [CrossRef] [PubMed]
- Mapelli, P.; Partelli, S.; Salgarello, M.; Doraku, J.; Pasetto, S.; Rancoita, P.M.V.; Muffatti, F.; Bettinardi, V.; Presotto, L.; Andreasi, V.; et al. Dual Tracer 68Ga-DOTATOC and 18F-FDG PET/Computed Tomography Ra-diomics in Pancreatic Neuroendocrine Neoplasms: An Endearing Tool for Preoperative Risk Assessment. Nucl. Med. Commun. 2020, 41, 896–905. [Google Scholar]
- Klimov, S.; Xue, Y.; Gertych, A.; Graham, R.P.; Jiang, Y.; Bhattarai, S.; Pandol, S.J.; Rakha, E.A.; Reid, M.D.; Aneja, R. Pre-dicting Metastasis Risk in Pancreatic Neuroendocrine Tumors Using Deep Learning Image Analysis. Front. Oncol. 2021, 10, 3336. [Google Scholar]
- Tang, T.; Liang, T.-B.; Zhang, Q.; Guo, C.; Zhang, X.; Lao, M.; Shen, Y.; Xiao, W.; Ying, S.; Sun, K.; et al. Development of a Novel Multiparametric MRI Radiomic Nomogram for Preoperative Evaluation of Early Recurrence in Resectable Pancreatic Cancer. J. Magn. Reson. Imaging 2019, 52, 231–245. [Google Scholar] [CrossRef]
- Bian, Y.; Guo, S.; Jiang, H.; Gao, S.; Shao, C.; Cao, K.; Fang, X.; Li, J.; Wang, L.; Hua, W.; et al. Relationship Between Radiomics and Risk of Lymph Node Metastasis in Pancreatic Ductal Adenocarcinoma. Pancreas 2019, 48, 1195–1203. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Gu, Q.; Hu, X.; Tan, X.; Liu, J.; Xie, A.; Huang, F. Applying a radiomics-based strategy to preoperatively predict lymph node metastasis in the resectable pancreatic ductal adenocarcinoma. J. X-ray Sci. Technol. 2020, 28, 1113–1121. [Google Scholar] [CrossRef]
- Bian, Y.; Jiang, H.; Ma, C.; Cao, K.; Fang, X.; Li, J.; Wang, L.; Zheng, J.; Lu, J. Performance of CT-Based Radiomics in Diagnosis of Superior Mesenteric Vein Resection Margin in Patients with Pancreatic Head Cancer. Abdom. Radiol. 2020, 45, 759–773. [Google Scholar]
- Hui, B.; Qiu, J.-J.; Liu, J.-H.; Ke, N.-W. Identification of Pancreaticoduodenectomy Resection for Pancreatic Head Adenocarcinoma: A Preliminary Study of Radiomics. Comput. Math. Methods Med. 2020, 2020, 1–12. [Google Scholar]
- Zhang, W.; Cai, W.; He, B.; Xiang, N.; Fang, C.; Jia, F. A Radiomics-Based Formula for the Preoperative Prediction of Postop-erative Pancreatic Fistula in Patients with Pancreaticoduodenectomy. Cancer Manag. Res. 2018, 10, 6469–6478. [Google Scholar] [PubMed] [Green Version]
- Li, J.; Shi, Z.; Liu, F.; Fang, X.; Cao, K.; Meng, Y.; Zhang, H.; Yu, J.; Feng, X.; Li, Q.; et al. XGBoost Classifier Based on Computed Tomography Radiomics for Prediction of Tumor-Infiltrating CD8+ T-Cells in Patients with Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2021, 11, 1714. [Google Scholar] [CrossRef]
- Bian, Y.; Liu, Y.F.; Jiang, H.; Meng, Y.; Liu, F.; Cao, K.; Zhang, H.; Fang, X.; Li, J.; Yu, J.; et al. Machine Learning for MRI Radiomics: A Study Predicting Tumor-Infiltrating Lymphocytes in Patients with Pancreatic Ductal Adenocarcinoma. Abdom. Radiol. 2021, 46, 4800–4816. [Google Scholar]
- Cassinotto, C.; Chong, J.; Zogopoulos, G.; Reinhold, C.; Chiche, L.; Lafourcade, J.-P.; Cuggia, A.; Terrebonne, E.; Dohan, A.; Gallix, B. Resectable Pancreatic Adenocarcinoma: Role of CT Quantitative Imaging Biomarkers for Predicting Pathology and Patient Outcomes. Eur. J. Radiol. 2017, 90, 152–158. [Google Scholar]
- Eilaghi, A.; Baig, S.; Zhang, Y.; Zhang, J.; Karanicolas, P.; Gallinger, S.; Khalvati, F.; Haider, M.A. CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma—A quantitative analysis. BMC Med Imaging 2017, 17, 38. [Google Scholar] [CrossRef]
- Shi, H.; Wei, Y.; Cheng, S.; Lu, Z.; Zhang, K.; Jiang, K.; Xu, Q. Survival Prediction after Upfront Surgery in Patients with Pancreatic Ductal Adenocarcinoma: Radiomic, Clinic-Pathologic and Body Composition Analysis. Pancreatology 2021, 21, 731–737. [Google Scholar]
- Yao, J.; Shi, Y.; Cao, K.; Lu, L.; Lu, J.; Song, Q.; Jin, G.; Xiao, J.; Hou, Y.; Zhang, L. DeepPrognosis: Preoperative Prediction of Pancreatic Cancer Survival and Surgical Margin via Comprehensive Understanding of Dynamic Contrast-Enhanced CT Imaging and Tumor-Vascular Contact Parsing. Med. Image Anal. 2021, 73, 102150. [Google Scholar]
- Cozzi, L.; Comito, T.; Fogliata, A.; Franzese, C.; Franceschini, D.; Bonifacio, C.; Tozzi, A.; Di Brina, L.; Clerici, E.; Tomatis, S.; et al. Computed Tomography Based Radiomic Signature as Predictive of Survival and Local Control after Stereotactic Body Radiation Thera-py in Pancreatic Carcinoma. PLoS ONE 2019, 14, e0210758. [Google Scholar]
- Steinacker, J.P.; Steinacker-Stanescu, N.; Ettrich, T.; Kornmann, M.; Kneer, K.; Beer, A.; Beer, M.; Schmidt, S.A. Computed Tomography-Based Tumor Heterogeneity Analysis Reveals Differences in a Cohort with Advanced Pancreatic Carcinoma under Palliative Chemotherapy. Visc. Med. 2020, 37, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Nasief, H.; Zheng, C.; Schott, D.; Hall, W.; Tsai, S.; Erickson, B.; Allen Li, X. A Machine Learning Based Delta-Radiomics Pro-cess for Early Prediction of Treatment Response of Pancreatic Cancer. Precis. Oncol. 2019, 3, 1–10. [Google Scholar]
- Nasief, H.; Hall, W.; Zheng, C.; Tsai, S.; Wang, L.; Erickson, B.; Li, X.A. Improving Treatment Response Prediction for Chemoradiation Therapy of Pancreatic Cancer Using a Combination of Delta-Radiomics and the Clinical Biomarker CA19-9. Front. Oncol. 2020, 9, 1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.-F.; Han, Y.-Q.; Lu, J.; Wei, J.-W.; Guo, J.-H.; Zhu, H.-D.; Huang, M.; Ji, J.-S.; Lv, W.-F.; Chen, L.; et al. Radiomics Facilitates Candidate Selection for Irradiation Stents Among Patients with Unresectable Pancreatic Cancer. Front. Oncol. 2019, 9, 973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Ding, Y.; Yu, Y.; Liu, K.; Rao, S.; Ge, Y.; Zeng, M. Whole-tumour evaluation with MRI and radiomics features to predict the efficacy of S-1 for adjuvant chemotherapy in postoperative pancreatic cancer patients: A pilot study. BMC Med. Imaging 2021, 21, 75. [Google Scholar] [CrossRef]
- Borhani, A.A.; Dewan, R.; Furlan, A.; Seiser, N.; Zureikat, A.H.; Singhi, A.D.; Boone, B.; Bahary, N.; Hogg, M.E.; Lotze, M.; et al. Assessment of Response to Neoadjuvant Therapy Using CT Texture Analysis in Patients with Resectable and Borderline Resectable Pancreatic Ductal Adenocarcinoma. Am. J. Roentgenol. 2020, 214, 362–369. [Google Scholar]
- Watson, M.D.; Baimas-George, M.R.; Murphy, K.J.; Pickens, R.C.; Iannitti, D.A.; Martinie, J.B.; Baker, E.H.; Vrochides, D.; Ocuin, L.M. Pure and Hybrid Deep Learning Models Can Predict Pathologic Tumor Response to Neoadjuvant Therapy in Pancreatic Adenocarcinoma: A Pilot Study. Am. Surg. 2020, 87, 1901–1909. [Google Scholar] [CrossRef]
- Chen, X.; Oshima, K.; Schott, D.; Wu, H.; Hall, W.; Song, Y.; Tao, Y.; Li, D.; Zheng, C.; Knechtges, P.; et al. Assessment of treatment response during chemoradiation therapy for pancreatic cancer based on quantitative radiomic analysis of daily CTs: An exploratory study. PLoS ONE 2017, 12, e0178961. [Google Scholar] [CrossRef] [Green Version]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging imaging and genomics. Abdom. Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef] [Green Version]
- McGovern, J.M.; Singhi, A.D.; Borhani, A.A.; Furlan, A.; McGrath, K.M.; Zeh, H.J.; Bahary, N.; Dasyam, A.K. CT Radi-ogenomic Characterization of the Alternative Lengthening of Telomeres Phenotype in Pancreatic Neuroendocrine Tumors. Am. J. Roentgenol. 2018, 211, 1020–1025. [Google Scholar]
- Gao, J.; Chen, X.; Li, X.; Miao, F.; Fang, W.; Li, B.; Qian, X.; Lin, X. Differentiating TP53 Mutation Status in Pancreatic Ductal Adenocarcinoma Using Multiparametric MRI-Derived Radiomics. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Katabathina, V.S.; Marji, H.; Khanna, L.; Ramani, N.; Yedururi, S.; Dasyam, A.; Menias, C.O.; Prasad, S.R. Decoding Genes: Current Update on Radiogenomics of Select Abdominal Malignancies. RadioGraphics 2020, 40, 1600–1626. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.; Baine, M.; Wisnoskie, S.; Bennion, N.; Zheng, D.; Yu, L.; Dalal, V.; Hollingsworth, M.A.; Lin, C.; Zheng, D. Effects of interobserver and interdisciplinary segmentation variabilities on CT-based radiomics for pancreatic cancer. Sci. Rep. 2021, 11, 16328. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, R.; Perrin, T.; Chakraborty, J.; Chou, J.F.; Horvat, N.; Koszalka, M.A.; Midya, A.; Gonen, M.; Allen, P.; Jarnagin, W.R.; et al. Radiomic feature reproducibility in contrast-enhanced CT of the pancreas is affected by variabilities in scan parameters and manual segmentation. Eur. Radiol. 2019, 30, 195–205. [Google Scholar] [CrossRef]
- Zhou, X. Automatic Segmentation of Multiple Organs on 3D CT Images by Using Deep Learning Approaches. Deep. Learn. Med Image Anal. 2020, 1213, 135–147. [Google Scholar] [CrossRef]
- Yang, G.; Ye, Q.; Xia, J. Unbox the black-box for the medical explainable AI via multi-modal and multi-centre data fusion: A mini-review, two showcases and beyond. Inf. Fusion 2021, 77, 29–52. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Lambin, P.; Woodruff, H. Radiomics Quality Score—RQS. Available online: https://www.radiomics.world/rqs (accessed on 8 July 2021).
- Rhee, H.; Park, M.-S. The Role of Imaging in Current Treatment Strategies for Pancreatic Adenocarcinoma. Korean J. Radiol. 2021, 22, 23–40. [Google Scholar] [CrossRef]


| Reference | Image | Software | Endpoints | Segmentation Process (Number of Readers) | Sample Size (Training + Validation) | Number of Features Extracted | Results |
|---|---|---|---|---|---|---|---|
| Attiyeh et al. [36] | CT | In-house software in MATLAB | BD-IPMN risk | manual (1) | 103 (10-fold cross-validation) | 255 | AUC = 0.79 for radiomics + clinical model vs. AUC = 0.67 for clinical model. |
| Chakraborty et al. [22] | CT | In-house software in MATLAB | BD-IPMN risk | manual (1) | 103 (10-fold cross-validation) | 150 | AUC = 0.77 for radiomics model and AUC = 0.81 for combined radiomics and clinical model. |
| Cheng et al. [29] | CT and MRI | ITK-SNAP software and Artificial Intelligence Kit software | predicting the malignant potential of intraductal papillary mucinous neoplasms (IPMNs) | manual (2) | 60 | 1037 | MRI radiomics models achieved improved AUCs (0.879 with LR and 0.940 with SVM, respectively), than that of CT radiomics models (0.811 with LR and 0.864 with SVM, respectively). All radiomics models provided better predictive performance than the clinical and imaging model (AUC = 0.764). |
| Cui et al. [26] | MRI | MITK software | Low vs. high-grade in BD-IPMNs | manual (2) | 103 + 48/51 (validation1/validation2) | 328 | Radiomics model: AUC = 0.836 (training); AUC = 0.811 (validation1); AUC = 0.822 (validation 2). Radiomics + clinical model: AUC = 0.903 (training); AUC = 0.884 (validation1); AUC = 0.876 (validation 2). |
| D‘Onofrio et al. [30] | MRI | MevisLasb and MATLAB | Identification and classification of IPMNs | manual (1) | 91 | <20 | Entropy of the ADC map was found to correlate with tumor dysplasia (p = 0.034, AUC = 0.729) |
| Hanania et al. [17] | CT | IBEX | High-grade vs. low-grade IPMNs | Manual (2) | 53 | 360 | Best univariate AUC = 0.82 |
| Harrington et al. [23] | CT | In-house software in MATLAB | IPMN risk | manual (1) | 33 | <20 | AUC = 0.74 (cyst fluid inflammatory markers model) vs. AUC = 0.83 (radiomics model) vs. AUC = 0.91 (tumor-associated neutrophils model) |
| Huang et al. (2021) [32] | CT | Pyradiomics | Invasiveness of SPN | Manual (2) | 85 | 1316 | Best AUC = 0.914 on 3D-arterial model (compared vs. 2D and venous) |
| Polk et al. [28] | CT | Healthmyne | Malignancy of IPMNs | semi-automatic (1, Healthmyne software) | 51 (5-fold cross-validation) | 39 | AUC = 0.87 (arterial model) vs. AUC = 0.83 (venous model) vs. AUC = 0.90 (combined) |
| Tobaly et al. [18] | CT | Pyradiomics | Differentiating IPMN grades | Manual (1) | 296 + 112 | 107 | AUC = 0.84 in training set and AUC = 0.71 in validation |
| Wei et al. [20] | CT | unknown | Computer-aided diagnosis of SCN | Manual (2) | 200 + 60 | 385 | AUC = 0.767 in training and AUC = 0.837 in validation |
| Xie et al. [21] | CT | In-house algorithm in MATLAB | Differentiating MCN vs. MaSCA | Manual (1) | 57 | 1942 | AUC = 0.989 (radiomics model) vs. AUC = 0.775 (radiological model) vs. AUC = 0.994 (combined model) on bootstrapping |
| Xie et al. [27] | CT | Pyradiomics | MCN vs. ASCN | semi-manually (1, 3D Slicer) | 216 (10-fold cross-validation) | 764 | Average AUC = 0.784 (radiomics model) vs. AUC = 0.734 (clinical model) |
| Yang et al. [37] | CT | LifeX | Differentiating SCA vs. MCA | manual (2) | 78 (4:1) | unknown | Slice thickness = 2 mm: AUC = 0.77 in training and AUC = 0.66 in validation; Slice thickness = 5 mm: AUC = 0.72 in training and AUC = 0.75 in validation |
| Reference | Image | Software | Endpoints | Sample Size (Training + Validation) | Results |
|---|---|---|---|---|---|
| Abel et al. [44] | CT | Two-step nnU-Net architecture | Detection of PCL | 221 (5-fold cross validation) | Mean sensitivity = 78.8% (87.8% for cysts ≥220 mm3 and 96.2% for lesions in distal pancreas) |
| Dmitriev et al. [41] | CT | CNN | Classification of 4 types of cysts: IPMN, MCN, SCA, SPN | 134 (10-fold cross validation) | Accuracy = 83.6% for the ensemble classifier (RF + CNN) |
| Luo et al. [40] | CT | CNN (ResNet50) | PNEN grading | 93 (8-fold cross validation) + 19 (independent testing set) | AUC = 0.81 (validation) AUC = 0.82 (testing) |
| Nguon et al. [45] | EUS | CNN using ResNet50 | MCNs vs. SCNs | 89 + 20 (holdout validation) | AUC = 0.88 for the classification of pancreatic SCNs and MCNs |
| Watson et al. [34] | CT | CNN (LeNet architecture) | PCN malignancy | 18 + 9 | AUC = 0.966 in high-risk lesions |
| Yang et al. [46] | CT | MMRF-ResNet | MCNs vs. SCNs | 110 (80:20 total images) | AUC = 0.96 for the classification of pancreatic SCNs and MCNs |
| Song et al. [33] | CT | * Fusion model. In-house software (manual segmentation by two observers, 143 radiomic features) | panNEN post-surgical recurrence risk | 56 + 18 | Better validation performance on arterial models with AUC = 0.77 (radiomics/DL fusion models) and AUC = 0.56 (radiomics model), compared to venous. |
| Reference | Image | Software | Endpoints | Segmentation Process (Number of Readers) | Sample Size (Training + Validation) | Number of Features Extracted | Results |
|---|---|---|---|---|---|---|---|
| Benedetti et al. [48] | CT | In house with Matlab | Discriminating histopathologic characteristics of PNET | Manual (1) | 39 | 69 | Best AUC = 0.86 |
| Bevilacqua et al. [49] | PET/CT | In house with Matlab | Grade 1 vs. 2 primary PNET | Manual (1) | 25 + 26 (model A) 26 + 25 (model B) 51 (model C) | 60 | Best performance was achieved by model A test AUC = 0.90 |
| Bian et al. [50] | MRI | Pyradiomics | G1 vs. G2/3 grades in patients with PNETs | Manual (2) | 157 | 1409 | AUC = 0.775 |
| Bian et al. [51] | MRI | Pyradiomics | PNET grades | Manual (1) | 97 + 42 | 3328 | AUC = 0.851 (training) AUC = 0.736 (validation) |
| Canellas et al. [52] | CT | TexRAD | Differentiating PNET grades | Manual (2) | 101 | 36 | Accuracy of 79.3% for differentiating grade1 vs. grades 2/3. |
| Chang et al. [53] | CT | IBEX | Histological grades of PDAC | Manual (2) | 151 + 150 (local) +100 (external validation) | 1452 | AUCs = 0.961 (training), AUC = 0.910 (local validation), and AUC = 0.770 (external validation) |
| Chen et al. [54] | CT | Pyradiomics | Differentiating PDAC from normal pancreas | Manual (2) | 915 + 200 (local test) + 264 (external test) | 88 | AUC = 0.98 (local test) AUC = 0.91 (external test) |
| Chu et al. [55] | CT | Pyradiomics | Differentiating PDAC from normal pancreas | Manual (3) | 255 + 125 | 478 | AUC = 0.999 |
| Deng et al. [56] | MRI | IBEX | DifferentiatingPDAC and MFCP lesions | Manual (2) | 64 + 55 | 410 | AUCs for the T1WI, T2WI, A and, P and clinical models were 0.893, 0.911, 0.958, 0.997 and 0.516 in the primary cohort, and 0.882, 0.902, 0.920, 0.962 and 0.649 in the validation cohort. |
| Gu et al. [57] | MRI | Artificial Intelligence Kit | SPN vs. differential diseases (PDAC, NET, and cystadenoma) | manual (2) | 48 + 113 | 2376 | In validation, AUC = 0.853 for T2 (best performing single sequence), AUC = 0.925 for multi-parametric MRI radiomics model, and AUC = 0.962 for radiomics + clinical model. |
| Li et al. [58] | CT | Fire Voxel | Atypical PNET vs. PDAC | Manual (2) | 75 | <20 | Best AUC = 0.887 |
| Linning et al. [59] | CT | In house with Matlab | PDAC vs. autoimmune pancreatitis | Manual (2) | 96 (5-fold cross validation) | 1160 | AUC = 0.977 |
| Liu et al. [60] | PET/CT | Pyradiomics | PDAC vs. autoimmune pancreatitis | Manual (2) | 112 (10-fold cross validation) | 502 | AUC= 0.967 |
| Liu et al. [61] | CT and MRI | Pyradiomics | PNET grades | Manual (2) | 82 + 41 | 1209 | AUC = 0.92 (training) AUC = 0.85 (validation) |
| Park et al. [62] | CT | Pyradiomics | PDAC vs. autoimmune pancreatitis | Manual (4) | 120 + 62 | 431 | AUC = 0.975 |
| Reinert et al. [63] | CT | Pyradiomics | Differentiating PDAC from PanNEN | Manual (1) | 95 | 92 | 8 features highly significant (p < 0.005) |
| Ren et al. [64] | CT | Analysis Kit software | Pancreatic adenosquamous carcinoma vs. PDAC | Manual (1) | 112 7:3 ratio | 792 | Average AUC of 0.82 |
| Song et al. [65] | MRI | Pyradiomics | Differentiating NF-PNET and SPN | Manual (2) | 79 (7:3 ratio) | 396 | AUC = 0.978 (radiomics) and AUC = 0.965 (radiomics + clinical) in the training set AUC = 0.907 (radiomics) and AUC = 0.920 (radiomics + clinical) in the validation set |
| Xing et al. [66] | PET/CT | Pyradiomics | Pathological grades in PDAC | Manual (2) | 99 + 50 | about 3000 | AUC o = 0.994 (training) AUC = 0.921 (validation) |
| Zhang et al. [67] | CT | LifeX | Pathological grades of PNETs | Manual (3) | 82 3:1 ratio | 40 | AUC = 0.82 (G1 vs. G2), 0.70 (G2 vs. G3), and 0.85 (G1 vs. G3), respectively |
| Zhao et al. [68] | CT | In house with Matlab | Grade 1 vs. 2 in PNET | Manual (2) | 59 + 40 | 585 | AUC = 0.968 (training) AUC= 0.876 (validation) |
| Reference | Image | Software | Endpoints | Sample Size (Training + Validation) | Results |
|---|---|---|---|---|---|
| Chu et al. [69] | CT | Deeply supervised nets with encoder-decoder architecture | PDAC detection | 456 | Sensitivity = 94.1%, specificity = 98.5% |
| Liu et al. [70] | CT | CNN | Differentiating pancreatic cancer vs. normal pancreas | 295 + 691 (local test 1 + local test 2 + external test) | AUC = 0.997 (local test 1) AUC = 0.999 (local test 2) AUC = 0.920 (external test) |
| Ozkan et al. [71] | EUS | ANN with Relief-F feature reduction method | Pancreatic cancer diagnosis for different age groups | 260 + 72 | Age groups in years: <40, 40–60, >60: accuracy = 92%, 88.5%, 91.7%, respectively all age groups: accuracy = 87.5% |
| Săftoiu et al. [72] | EUS | ANN (MLP) | Differential diagnosis of chronic pancreatitis and pancreatic cancer | 68 (10-fold cross validation) | Benign vs. malignant pancreatic lesions: AUC = 0.957 Chronic pancreatitis vs. pancreatic cancer: AUC = 0.965 |
| Săftoiu et al. [73] | EUS | ANN (MLP) | Diagnosis of focal pancreatic masses | 258 (10-fold cross validation) | Average AUC = 0.94 over 100 runs of a complete cross-validation cycle |
| Si et al. [74] | CT | CNN ResNet18 (pancreas location), U-Net32 (pancreas segmentation), ResNet34 (pancreatic tumor diagnosis) | Fully automated diagnosis of pancreatic tumors | 319 + 347 | AUC = 0.871 on testing for detection of all tumor types |
| Tonozuka et al. [75] | EUS | CNN | PDAC detection | 92 (10-fold cross validation) + 47 | AUC = 0.924 (cross validation) AUC = 940 (test) |
| Zhang et al. [76] | CT | Faster R-CNN combined with Feature Pyramid Network for feature extraction | Pancreatic tumor detection | 2650 + 240 (images) | AUC = 0.946 |
| Reference | Image | Software | Endpoints | Segmentation Process (Number of Readers) | Sample Size (Training + Validation) | Number of Features Extracted | Results |
|---|---|---|---|---|---|---|---|
| Bian et al. [103] | CT | Pyradiomics | Lymph node metastasis in PDAC | Manual (2) | 225 (10-fold cross validation) | 1029 | Multivariate p < 0.0001 |
| Bian et al. [105] | CT | Pyradiomics | R0 vs. R1 margin in pancreatic head cancer | Manual (2) | 181 (10-fold cross validation) | 1029 | AUC = 0.750 |
| Bian et al. [109] | MRI | Pyradiomics | Tumor-infiltrating lymphocytes in patients with PDAC | Manual (2) | 116 + 40 | 1409 | training AUC = 0.86 and validation sets AUC = 0.79 |
| Cassinottoet al. [110] | CT | TexRAD | Disease-free survival in patients with resectable PDAC | Manual (1) | 99 | <20 (texture) | AUC 0.71 |
| Cen et al. [84] | CT | Analysis Kit software | Stage I-II vs. III-IV PDAC and predict overall survival | Manual (2) | 94 + 41 | 384 | Training cohort AUC = 0.940 Validation cohort AUC = 0.912 |
| Cheng et al. [80] | CT | TexRAD | Progression-free survival and overall survival in patients with unresectable PDAC | Manual (1) | 41 | <20 (texture) | AUC = 0.756 |
| Cusumano et al. [83] | MRI | MODDICOM software | One-year local control in patients with locally advanced pancreatic cancer | Manual (2) | 35 (5-fold cross validation) | 368 radiomic features and 276 delta features | AUC = 0.78 |
| D’Onofrio et al. [93] | CT | In house with unknown software | Metastatic vs. non-metastatic PDAC | Manual (1) | 288 | <20 | Significant univariate features identified: size, arterial index, perfusion index, and permeability index (p < 0.05). |
| Eilaghi et al. [111] | CT | In house with Matlab | Overall survival for PDAC after surgical resection | Semi-automatic (1, in-house ProCanVAS) | 30 | <20 | Max AUC = 0.716 in univariate |
| Hang et al. [89] | CT | LifeX | Overall survival for pancreatic cancer with liver metastases | Manual (1) | 39 | 36 | Nomogram showed good discriminative ability (CI = 0.754). |
| Hui et al. [106] | CT | Rbio2.8 | R0 or R1 margin in pancreatic head adenocarcinoma | Manual (2) | 86 (leave-one-out cross validation) | 23 | AUC = 0.861 |
| Kaissis et al. [95] | MRI | Pyradiomics | Survival and tumor subtype in PDAC | Manual (2) | 102 (10-fold nested cross validation) + 30 | 1474 | AUC = 0.93 in cross-validation AUC = 0.90 in independent validation |
| Khalvati et al. [86] | CT | Pyradiomics | Prognostic value of CT-derived radiomic features for resectable PDAC | Manual (2) | 30 + 68 | 410 | Validation cohort with p-value of 0.047 |
| Kim et al. [85] | CT | In house with unknown software | predict prognosis after curative resection in pancreatic cancer | Manual (1) | 116 | <20 (GLRLM) | One feature with p = 0.025 for survival |
| Li et al. [82] | CT | Pyradiomics | Lymph node metastasis | Manual (2) | 118 + 41 | 2041 | Best AUC = 0.811 |
| Li et al. [108] | CT | Pyradiomics | CD8+ tumor-infiltrating lymphocyte expression levels in patients with PDAC | Manual (2) | 137 + 47 | 1409 | Training set AUC = 0.75 and validation set AUC = 0.67 |
| Liu et al. [104] | CT | Pyradiomics | Lymph node metastasis in resectable PDAC | Manual (2) | 85 | 1124 | AUC = 0.841 (radiomics) vs. AUC = 0.682 (conventional) |
| Mapelli et al. [100] | PET/CT | Chang-Gung Image Texture Analysis software package | PanNEN risks | Automatic with SUV thresholding (40% of SUVmax) | 61 | 9 | Four principal components extracted: PC1 correlated with all 18F-FDG variables, while PC2, PC3 and PC4 with 68Ga-DOTATOC variables. PC1 could predict angioinvasion (p = 0.0222); PC4 could predict lymph nodal involvement (p = 0.0151). All PCs except PC4 could predict tumor dimension |
| Mapelli et al. [94] | PET/CT | Chang-Gung Image Texture Analysis software package | PanNEN risks | Automatic with SUV thresholding (40% of SUVmax) | 83 | 9 | Individual parameters evaluated for various clinical risk endpoints |
| Mori et al. [90] | PET | Spaarc Pipeline for Automated Analysis and Radiomics Computing (SPAARC) | Distant-relapse-free-survival after radio-chemotherapy for locally advanced pancreatic cancer | Semi-automatic (gradient based, PET-Edge, MIM) | 116 + 60 | 198 | Training cohort p = 0.002 and validation cohort p = 0.03. |
| Salinas-Miranda et al. [91] | CT | Pyradiomics | Overall survival and time to progression; validate radiomic features developed in resectable PDAC on a test set of patients with unresectable PDAC undergoing chemotherapy | Manual (1) | 0 + 108 | 2 previously developed features | One feature remained significant with a HR = 1.27 for overall survival and a HR of 1.25 for time to progression |
| Shi et al. [112] | CT | ITK-SNAP software and Artificial Intelligent Kit | Survival after upfront surgery in patients with PDAC | Manual (2) | 210 + 89 | 792 | CI = 0.74 in the training set and CI = 0.73 in the validation set. |
| Tang et al. [102] | MRI | AK software | Early recurrence in resectable pancreatic cancer | Manual (2) | 123 + 54 (+126 external validation) | 328 | AUC = 0.871 (training cohort), AUC = 0.876 (internal validation cohort), and AUC = 0.846 (external validation cohort). |
| Toyama et al. [87] | PET | LifeX and machine learning algorithms | 1-year survival | Semi-automatic (2, with SUV thresholding at 40% of SUVmax) | 161 (10-fold cross validation on 138) | 42 | Best AUC = 0.720 |
| Xie et al. [88] | CT | Mazda | Survival in patients with resected PDAC | Manual (3) | 147 + 73 | 300 | AUC = 0.701 in training cohort AUC = 0.715 in validation cohort |
| Zhang et al. [107] | CT | Pyradiomics | Postoperative pancreatic fistula after pancreaticoduodenectomy | Manual (2) | 80 + 37 | 1219 | AUC = 0.825 in training cohort and AUC = 0.761 in validation cohort |
| Reference | Image | Software | Endpoints | Sample Size (Training + Validation) | Results |
|---|---|---|---|---|---|
| Gao et al. [99] | MRI | CNN combined with GAN for synthetic image generation | PNET grades | 96 (5-fold cross validation) + 10 | Micro-average AUC = 0.912 in internal validation set; Micro-average AUC = 0.845 in external validation set |
| Klimov et al. [101] | Whole-slide imaging of resected tissues | CNN for tissue annotation, 18 different ML models for metastasis prediction | Metastasis risk in PNET | 89 | Tissue annotation: per-tile accuracy > 95%, whole slide 79%; Metastasis prediction: hazard ratio 4.71 |
| Li et al. [92] | CT | Fusion model (70 conventional features and 256 deep convolutional features) Matlab | Survival time in PDAC | 111 (k-fold leave-one-out cross validation, k = 10, 20, 30, 40) | Average AUC = 0.90 |
| Yao et al.(2020) [96] | CT | * Fusion model. Pyradiomics, CNN (CE- convLSTM, combined with 3D-ResNet18 as the encoder) | PDAC survival and surgical margin | 205 (5-fold cross validation) | survival prediction: C-index = 0.705; resection margin prediction: balanced-accuracy = 0.736 |
| Yao et al. [113] | CT | CNN | Survival of primary resectable PDAC | 296 (4-fold nested cross validation) | 1-year overall survival: AUC = 0.684; 2-year overall survival: AUC = 0.689 |
| Zhang et al. [97] | CT | CNN-based transfer learning model | prognosis of overall survival in PDAC patients | 68 (5-fold cross validation) + 30 | AUC = 0.72 in training cohort; AUC = 0.81 in test cohort |
| Zhang et al. [98] | CT | * Fusion model. Pyradiomics. Random forest-based models trained from features extracted using traditional radiomics pipeline and transfer learning | Overall survival in PDAC | 68 (10-fold cross validation) + 30 | AUC = 0.84 in test cohort |
| Reference | Image | Software | Endpoints | Segmentation Process (Number of Readers) | Sample Size (Training + Validation) | Number of Features Extracted | Results |
|---|---|---|---|---|---|---|---|
| Borhani et al. [120] | CT | TexRAD | Histologic response to neoadjuvant CRT and disease-free survival in patients with potentially resectable PDAC | Manual (1) | 39 | <20 for each filter, 6 filters applied | Prognostic features identified for histological response (p < 0.05), biochemical response (p < 0.01) and disease-free survival (p = 0.001). |
| Chen et al. [122] | CT | In house with Matlab | Delta-radiomic change during CRT and pathology responses on 15 patients that undergone subsequent resections | Manual (1) | 20 | <20 | p = 0.046, 0.058, 0.042, and 0.12 for MCTN, SD, skewness and kurtosis, respectively. |
| Cozzi et al. [114] | CT | LifeX | Overall survival after stereotactic body radiation therapy | Manual (1) | 60 + 40 | 41 | AUC = 0.81 for the training set and AUC = 0.73 for the validation set |
| Liang et al. [119] | MRI | Pyradiomics | Efficacy of S-1 (oral antitumor agent) | Semi-automatic (2, a generic automatic segmentation algorithm based on a 3D domain using a prototype software, Radiomics, Siemens) | 31 + 15 | 110 | T1WI_NGTDM_Strength and tumor location are independent predictors of the efficacy of S-1 in the training cohort (p = 0.005 and 0.013), but marginal in the validation cohort (p = 0.073 and 0.050). |
| Nasief et al. [116] | CT | IBEX | Delta-radiomic change and overall progression in patients undergone neoadjuvant CRT | Manual (1) | 50 (leave-one-out cross validation) + 40 (external) | >1300 | Best AUC = 0.94 |
| Nasief et al. [117] | CT | IBEX | Delta-radiomic change and overall progression in patients undergone neoadjuvant CRT | Manual (1) | 24 | Over 1300 | The Cox proportional multivariate hazard analysis showed that a treatment related decrease in CA19-9 levels (p = 0.031) and delta radiomics (p = 0.001) were predictors of survival. |
| Parr et al. [81] | CT | Pyradiomics | Overall survival and locoregional recurrence following stereotactic body radiation | Manual (2) | 74 (3-fold cross validation) | 841 | Validation: Average CI of 0.66 (radiomics) vs. 0.54 (clinical) for survival; Average AUC of 0.78 (radiomics) vs. 0.66 (clinical) for recurrence. |
| Steinacker et al. [115] | CT | MintLesion | Overall progression in advanced pancreatic cancer treated with systemic therapy | Semi-automatic (1, mintLesion®.) | 13 | <20 | Two significant univariate features identified: mean positivity of pixel values (p = 0.030 for progression); kurtosis (p = 0.008 for time to local tumor spread and p = 0.017 for systemic progression). |
| Watson et al. [121] | CT | CNN (based onLeNet architecture) | Pathologic tumor response to neoadjuvant therapy in pancreatic adenocarcinoma | NA (deep learning) | 65 + 16 | NA (deep learning) | AUC = 0.738 (DL), AUC = 0.564 (CA19-9), and AUC = 0.785 (combined) |
| Zhou et al. [118] | CT | In house with Matlab | Candidate selection for irradiation stent placement among patients with unresectable pancreatic cancer with malignant biliary obstruction | Manual (2) | 74 + 32 | 620 | CI = 0.791 (radiomics + clinical) vs. CI = 0.673 (clinical) in the training set; CI = 0.779 (radiomics + clinical) vs. CI = 0.667 (clinical) in the validation groups |
| Attiyeh et al. [36] | CT | Matlab | CT imaging phenotypes and genetic and biological characteristics PDAC | Manual (1) | 35 | 255 | Radiomics associated with SMAD4 status and the number of genes altered |
| Gao et al. [125] | MRI | Pyradiomics | TP53 mutation status | Manual (2) | 57 | 558 2D and 994 3D features | AUC = 0.96 |
| Iwatate et al. [9] | CT | Pyradiomics | Genetic information | Manual (2) | 107 | 1037 | Radiogenomics-predicted p53 mutations associated with poor prognosis (p = 0.02), whereas the predicted abnormal expression of PD-L1 was not significant (p = 0.10). |
| Lim et al. [8] | PET | Chang-Gung Image Texture Analysis | KRAS, SMAD4, TP53, and CDKN2A mutation status | Semi-automatic (3, gradient based, PET-Edge, MIM) | 116 + 60 | 35 | Features identified that associated with KRAS and SMAD4 gene mutations, but not with TP53 and CDKN2A gene mutations. |
| McGovern et al. [124] | CT | Unknown | Predicting the ALT phenotype in PNET patients | Manual (2) | 121 | <20 | Univariate (p < 0.05) and multivariate features (p = 0.006) found. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Preuss, K.; Thach, N.; Liang, X.; Baine, M.; Chen, J.; Zhang, C.; Du, H.; Yu, H.; Lin, C.; Hollingsworth, M.A.; et al. Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications. Cancers 2022, 14, 1654. https://doi.org/10.3390/cancers14071654
Preuss K, Thach N, Liang X, Baine M, Chen J, Zhang C, Du H, Yu H, Lin C, Hollingsworth MA, et al. Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications. Cancers. 2022; 14(7):1654. https://doi.org/10.3390/cancers14071654
Chicago/Turabian StylePreuss, Kiersten, Nate Thach, Xiaoying Liang, Michael Baine, Justin Chen, Chi Zhang, Huijing Du, Hongfeng Yu, Chi Lin, Michael A. Hollingsworth, and et al. 2022. "Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications" Cancers 14, no. 7: 1654. https://doi.org/10.3390/cancers14071654
APA StylePreuss, K., Thach, N., Liang, X., Baine, M., Chen, J., Zhang, C., Du, H., Yu, H., Lin, C., Hollingsworth, M. A., & Zheng, D. (2022). Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications. Cancers, 14(7), 1654. https://doi.org/10.3390/cancers14071654







