Radiomics and Cancers
Share This Topical Collection
Editor
Topical Collection Information
Dear Colleagues,
Different radiological imaging methods are used in oncology to investigate different regions of the body for the detection of primary tumors and metastatic spread (i.e., for tumor staging). At present, imaging can also characterize several lesions and predict their histopathological features. Furthermore, imaging can predict tumor behavior and prognosis.
Radiomics represents a novel imaging analysis technique, consisting of the extraction of numerous quantitative features using automated or semi-automated software. Radiomics is based on the hypothesis that mineable data can be extracted from medical images and provide additional information on gene protein and tumor phenotype, which can then be used for patient care.
The purpose of the present collection is to analyze the possible relationships between different histopathological features and radiomics parameters in several malignancies.
Prof. Dr. Alexey Surov
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Cancers is an international peer-reviewed open access semimonthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 2900 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- radiomics
- imaging
- oncology
- histopathology
- survival
Published Papers (6 papers)
Open AccessReview
Prostate-Specific Membrane Antigen-Positron Emission Tomography-Guided Radiomics and Machine Learning in Prostate Carcinoma
by
Justine Maes, Simon Gesquière, Alex Maes, Mike Sathekge and Christophe Van de Wiele
Viewed by 1426
Abstract
Positron emission tomography (PET) using radiolabeled prostate-specific membrane antigen targeting PET-imaging agents has been increasingly used over the past decade for imaging and directing prostate carcinoma treatment. Here, we summarize the available literature data on radiomics and machine learning using these imaging agents
[...] Read more.
Positron emission tomography (PET) using radiolabeled prostate-specific membrane antigen targeting PET-imaging agents has been increasingly used over the past decade for imaging and directing prostate carcinoma treatment. Here, we summarize the available literature data on radiomics and machine learning using these imaging agents in prostate carcinoma. Gleason scores derived from biopsy and after resection are discordant in a large number of prostate carcinoma patients. Available studies suggest that radiomics and machine learning applied to PSMA-radioligand avid primary prostate carcinoma might be better performing than biopsy-based Gleason-scoring and could serve as an alternative for non-invasive GS characterization. Furthermore, it may allow for the prediction of biochemical recurrence with a net benefit for clinical utilization. Machine learning based on PET/CT radiomics features was also shown to be able to differentiate benign from malignant increased tracer uptake on PSMA-targeting radioligand PET/CT examinations, thus paving the way for a fully automated image reading in nuclear medicine. As for prediction to treatment outcome following
177Lu-PSMA therapy and overall survival, a limited number of studies have reported promising results on radiomics and machine learning applied to PSMA-targeting radioligand PET/CT images for this purpose. Its added value to clinical parameters warrants further exploration in larger datasets of patients.
Full article
Open AccessArticle
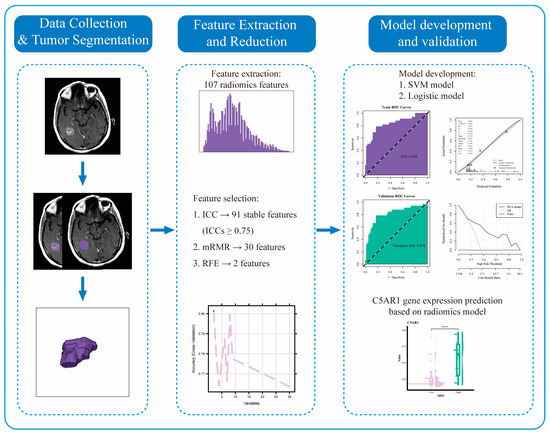
Radiomics Features on Magnetic Resonance Images Can Predict C5aR1 Expression Levels and Prognosis in High-Grade Glioma
by
Zijun Wu, Yuan Yang and Yunfei Zha
Cited by 3 | Viewed by 1767
Abstract
Background: The complement component C5a receptor 1 (C5aR1) regulates cancer immunity. This retrospective study aimed to assess its prognostic value in high-grade glioma (HGG) and predict C5aR1 expression using a radiomics approach. Methods: Among 298 patients with HGG, 182 with MRI data were
[...] Read more.
Background: The complement component C5a receptor 1 (C5aR1) regulates cancer immunity. This retrospective study aimed to assess its prognostic value in high-grade glioma (HGG) and predict C5aR1 expression using a radiomics approach. Methods: Among 298 patients with HGG, 182 with MRI data were randomly divided into training and test groups for radiomics analysis. We examined the association between C5aR1 expression and prognosis through Kaplan–Meier and Cox regression analyses. We used maximum relevance–minimum redundancy and recursive feature elimination algorithms for radiomics feature selection. We then built a support vector machine (SVM) and a logistic regression model, investigating their performances using receiver operating characteristic, calibration curves, and decision curves. Results: C5aR1 expression was elevated in HGG and was an independent prognostic factor (hazard ratio = 3.984, 95% CI: 2.834–5.607). Both models presented with >0.8 area under the curve values in the training and test datasets, indicating efficient discriminatory ability, with SVM performing marginally better. The radiomics score calculated using the SVM model correlated significantly with overall survival (
p < 0.01). Conclusions: Our results highlight C5aR1’s role in HGG development and prognosis, supporting its potential as a prognostic biomarker. Our radiomics model can noninvasively and effectively predict C5aR1 expression and patient prognosis in HGG.
Full article
►▼
Show Figures
Open AccessArticle
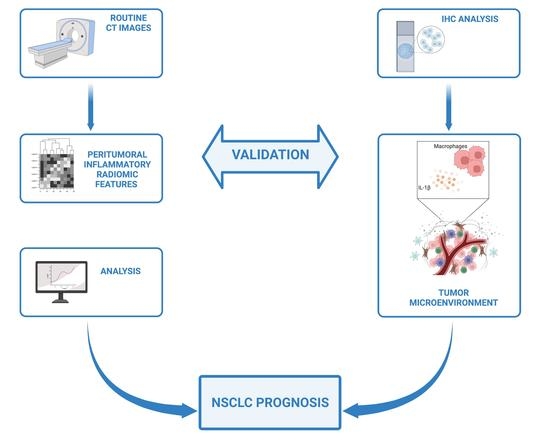
Inflammatory Microenvironment in Early Non-Small Cell Lung Cancer: Exploring the Predictive Value of Radiomics
by
Mariasole Perrone, Edoardo Raimondi, Matilde Costa, Gianluca Rasetto, Roberto Rizzati, Giovanni Lanza, Roberta Gafà, Giorgio Cavallesco, Nicola Tamburini, Pio Maniscalco, Maria Cristina Mantovani, Umberto Tebano, Manuela Coeli, Sonia Missiroli, Massimo Tilli, Paolo Pinton, Carlotta Giorgi and Francesco Fiorica
Cited by 8 | Viewed by 2640
Abstract
Patient prognosis is a critical consideration in the treatment decision-making process. Conventionally, patient outcome is related to tumor characteristics, the cancer spread, and the patients’ conditions. However, unexplained differences in survival time are often observed, even among patients with similar clinical and molecular
[...] Read more.
Patient prognosis is a critical consideration in the treatment decision-making process. Conventionally, patient outcome is related to tumor characteristics, the cancer spread, and the patients’ conditions. However, unexplained differences in survival time are often observed, even among patients with similar clinical and molecular tumor traits. This study investigated how inflammatory radiomic features can correlate with evidence-based biological analyses to provide translated value in assessing clinical outcomes in patients with NSCLC. We analyzed a group of 15 patients with stage I NSCLC who showed extremely different OS outcomes despite apparently harboring the same tumor characteristics. We thus analyzed the inflammatory levels in their tumor microenvironment (TME) either biologically or radiologically, focusing our attention on the NLRP3 cancer-dependent inflammasome pathway. We determined an NLRP3-dependent peritumoral inflammatory status correlated with the outcome of NSCLC patients, with markedly increased OS in those patients with a low rate of NLRP3 activation. We consistently extracted specific radiomic signatures that perfectly discriminated patients’ inflammatory levels and, therefore, their clinical outcomes. We developed and validated a radiomic model unleashing quantitative inflammatory features from CT images with an excellent performance to predict the evolution pattern of NSCLC tumors for a personalized and accelerated patient management in a non-invasive way.
Full article
►▼
Show Figures
Open AccessEditor’s ChoiceReview
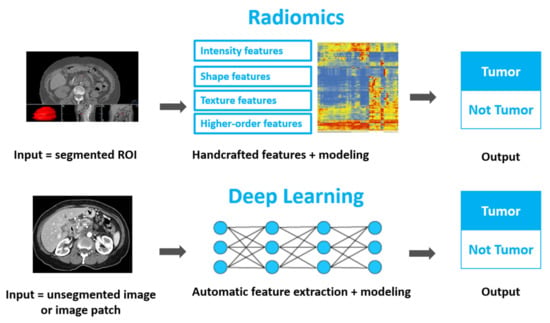
Using Quantitative Imaging for Personalized Medicine in Pancreatic Cancer: A Review of Radiomics and Deep Learning Applications
by
Kiersten Preuss, Nate Thach, Xiaoying Liang, Michael Baine, Justin Chen, Chi Zhang, Huijing Du, Hongfeng Yu, Chi Lin, Michael A. Hollingsworth and Dandan Zheng
Cited by 58 | Viewed by 10801
Abstract
As the most lethal major cancer, pancreatic cancer is a global healthcare challenge. Personalized medicine utilizing cutting-edge multi-omics data holds potential for major breakthroughs in tackling this critical problem. Radiomics and deep learning, two trendy quantitative imaging methods that take advantage of data
[...] Read more.
As the most lethal major cancer, pancreatic cancer is a global healthcare challenge. Personalized medicine utilizing cutting-edge multi-omics data holds potential for major breakthroughs in tackling this critical problem. Radiomics and deep learning, two trendy quantitative imaging methods that take advantage of data science and modern medical imaging, have shown increasing promise in advancing the precision management of pancreatic cancer via diagnosing of precursor diseases, early detection, accurate diagnosis, and treatment personalization and optimization. Radiomics employs manually-crafted features, while deep learning applies computer-generated automatic features. These two methods aim to mine hidden information in medical images that is missed by conventional radiology and gain insights by systematically comparing the quantitative image information across different patients in order to characterize unique imaging phenotypes. Both methods have been studied and applied in various pancreatic cancer clinical applications. In this review, we begin with an introduction to the clinical problems and the technology. After providing technical overviews of the two methods, this review focuses on the current progress of clinical applications in precancerous lesion diagnosis, pancreatic cancer detection and diagnosis, prognosis prediction, treatment stratification, and radiogenomics. The limitations of current studies and methods are discussed, along with future directions. With better standardization and optimization of the workflow from image acquisition to analysis and with larger and especially prospective high-quality datasets, radiomics and deep learning methods could show real hope in the battle against pancreatic cancer through big data-based high-precision personalization.
Full article
►▼
Show Figures
Open AccessArticle
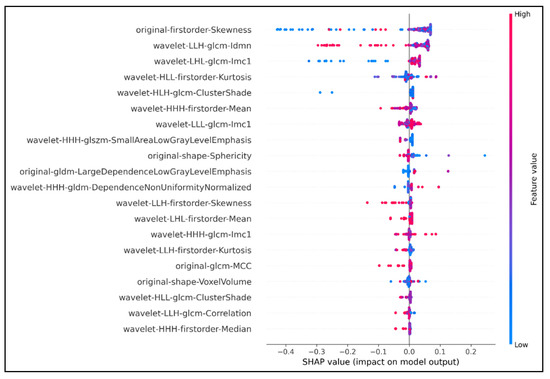
Development and Validation of an Efficient MRI Radiomics Signature for Improving the Predictive Performance of 1p/19q Co-Deletion in Lower-Grade Gliomas
by
Quang-Hien Kha, Viet-Huan Le, Truong Nguyen Khanh Hung and Nguyen Quoc Khanh Le
Cited by 41 | Viewed by 4873
Abstract
The prognosis and treatment plans for patients diagnosed with low-grade gliomas (LGGs) may significantly be improved if there is evidence of chromosome 1p/19q co-deletion mutation. Many studies proved that the codeletion status of 1p/19q enhances the sensitivity of the tumor to different types
[...] Read more.
The prognosis and treatment plans for patients diagnosed with low-grade gliomas (LGGs) may significantly be improved if there is evidence of chromosome 1p/19q co-deletion mutation. Many studies proved that the codeletion status of 1p/19q enhances the sensitivity of the tumor to different types of therapeutics. However, the current clinical gold standard of detecting this chromosomal mutation remains invasive and poses implicit risks to patients. Radiomics features derived from medical images have been used as a new approach for non-invasive diagnosis and clinical decisions. This study proposed an eXtreme Gradient Boosting (XGBoost)-based model to predict the 1p/19q codeletion status in a binary classification task. We trained our model on the public database extracted from The Cancer Imaging Archive (TCIA), including 159 LGG patients with 1p/19q co-deletion mutation status. The XGBoost was the baseline algorithm, and we combined the SHapley Additive exPlanations (SHAP) analysis to select the seven most optimal radiomics features to build the final predictive model. Our final model achieved an accuracy of 87% and 82.8% on the training set and external test set, respectively. With seven wavelet radiomics features, our XGBoost-based model can identify the 1p/19q codeletion status in LGG-diagnosed patients for better management and address the drawbacks of invasive gold-standard tests in clinical practice.
Full article
►▼
Show Figures
Open AccessArticle
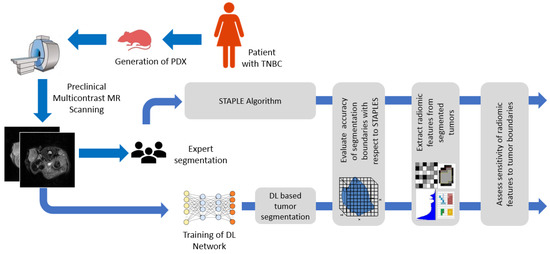
Deep Learning Segmentation of Triple-Negative Breast Cancer (TNBC) Patient Derived Tumor Xenograft (PDX) and Sensitivity of Radiomic Pipeline to Tumor Probability Boundary
by
Kaushik Dutta, Sudipta Roy, Timothy Daniel Whitehead, Jingqin Luo, Abhinav Kumar Jha, Shunqiang Li, James Dennis Quirk and Kooresh Isaac Shoghi
Cited by 30 | Viewed by 5059
Abstract
Preclinical magnetic resonance imaging (MRI) is a critical component in a co-clinical research pipeline. Importantly, segmentation of tumors in MRI is a necessary step in tumor phenotyping and assessment of response to therapy. However, manual segmentation is time-intensive and suffers from inter- and
[...] Read more.
Preclinical magnetic resonance imaging (MRI) is a critical component in a co-clinical research pipeline. Importantly, segmentation of tumors in MRI is a necessary step in tumor phenotyping and assessment of response to therapy. However, manual segmentation is time-intensive and suffers from inter- and intra- observer variability and lack of reproducibility. This study aimed to develop an automated pipeline for accurate localization and delineation of TNBC PDX tumors from preclinical T1w and T2w MR images using a deep learning (DL) algorithm and to assess the sensitivity of radiomic features to tumor boundaries. We tested five network architectures including U-Net, dense U-Net, Res-Net, recurrent residual UNet (R2UNet), and dense R2U-Net (D-R2UNet), which were compared against manual delineation by experts. To mitigate bias among multiple experts, the simultaneous truth and performance level estimation (STAPLE) algorithm was applied to create consensus maps. Performance metrics (F1-Score, recall, precision, and AUC) were used to assess the performance of the networks. Multi-contrast D-R2UNet performed best with F1-score = 0.948; however, all networks scored within 1–3% of each other. Radiomic features extracted from D-R2UNet were highly corelated to STAPLE-derived features with 67.13% of T1w and 53.15% of T2w exhibiting correlation ρ ≥ 0.9 (
p ≤ 0.05). D-R2UNet-extracted features exhibited better reproducibility relative to STAPLE with 86.71% of T1w and 69.93% of T2w features found to be highly reproducible (CCC ≥ 0.9,
p ≤ 0.05). Finally, 39.16% T1w and 13.9% T2w features were identified as insensitive to tumor boundary perturbations (Spearman correlation (−0.4 ≤ ρ ≤ 0.4). We developed a highly reproducible DL algorithm to circumvent manual segmentation of T1w and T2w MR images and identified sensitivity of radiomic features to tumor boundaries.
Full article
►▼
Show Figures