A Search for Dual Action HIV-1 Reverse Transcriptase, Bacterial RNA Polymerase Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Rationale
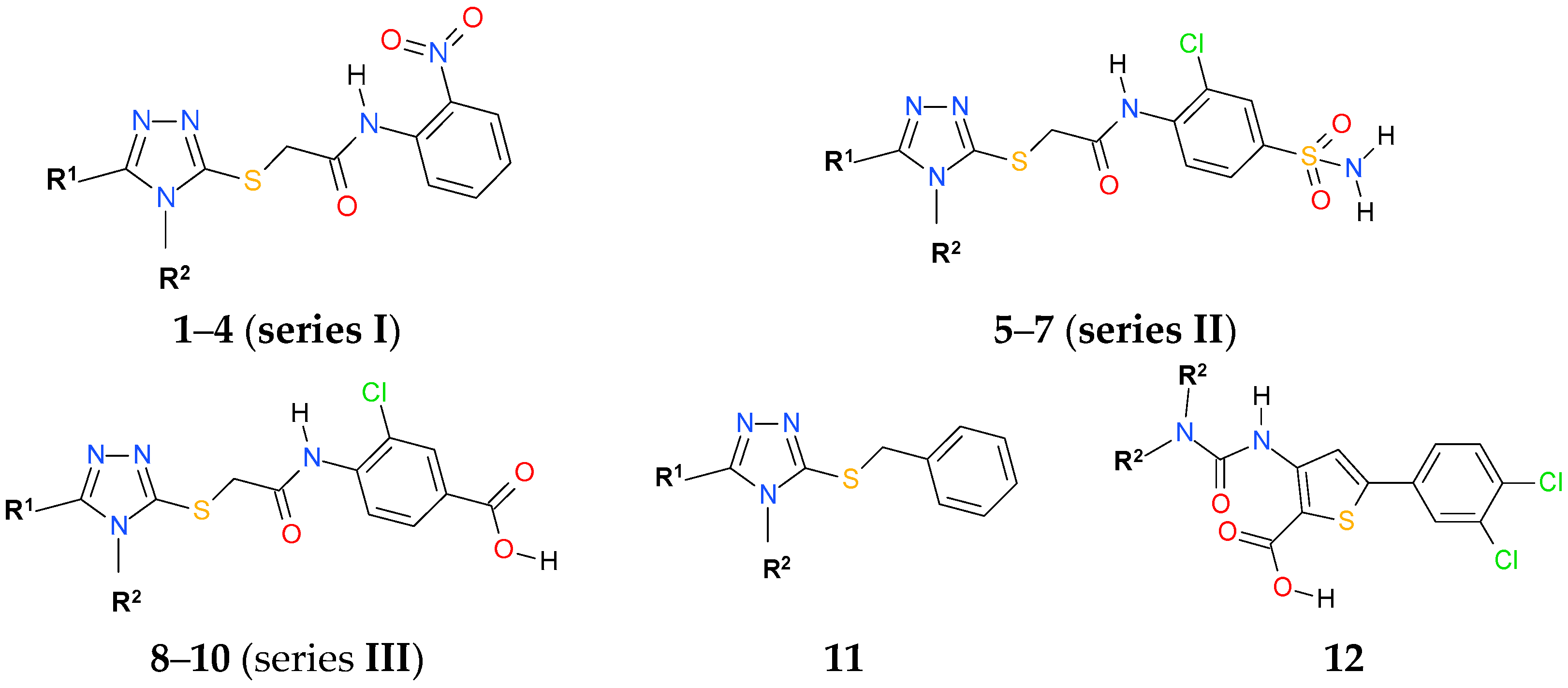
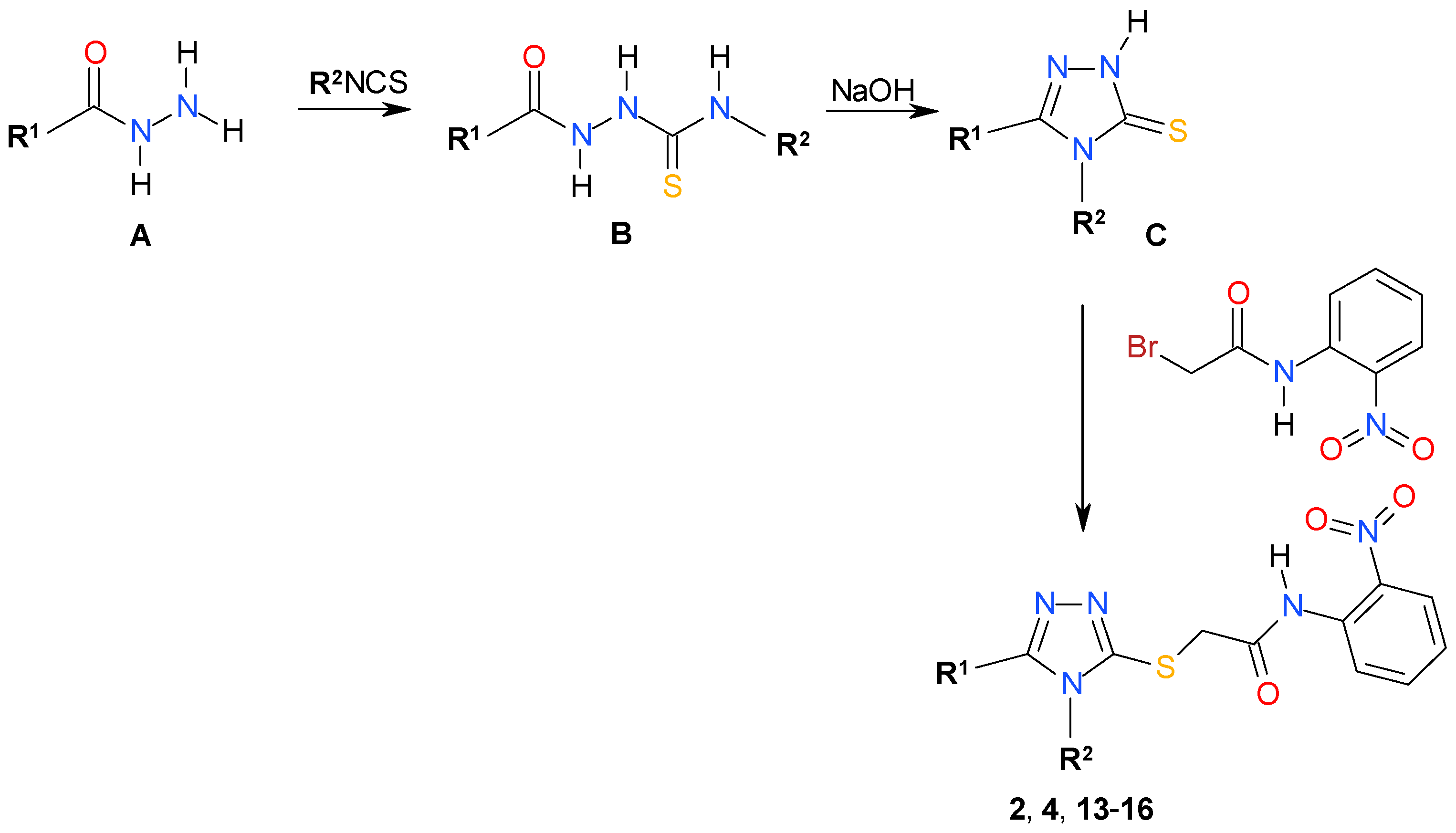
2.2. Chemistry
2.3. Antimicrobial Assay
3. Materials and Methods
3.1. Chemistry
3.1.1. General Procedure for Synthesis of Thiosemicarbazides B
3.1.2. General Procedure for Synthesis of Triazoles C
3.1.3. Synthesis of 2-Bromo-N-(2-nitrophenyl)acetamide
3.1.4. General Procedure for Synthesis of the Triazoles (2, 4, 13–16)
3.2. Antimicrobial Assay
3.3. Computational Details
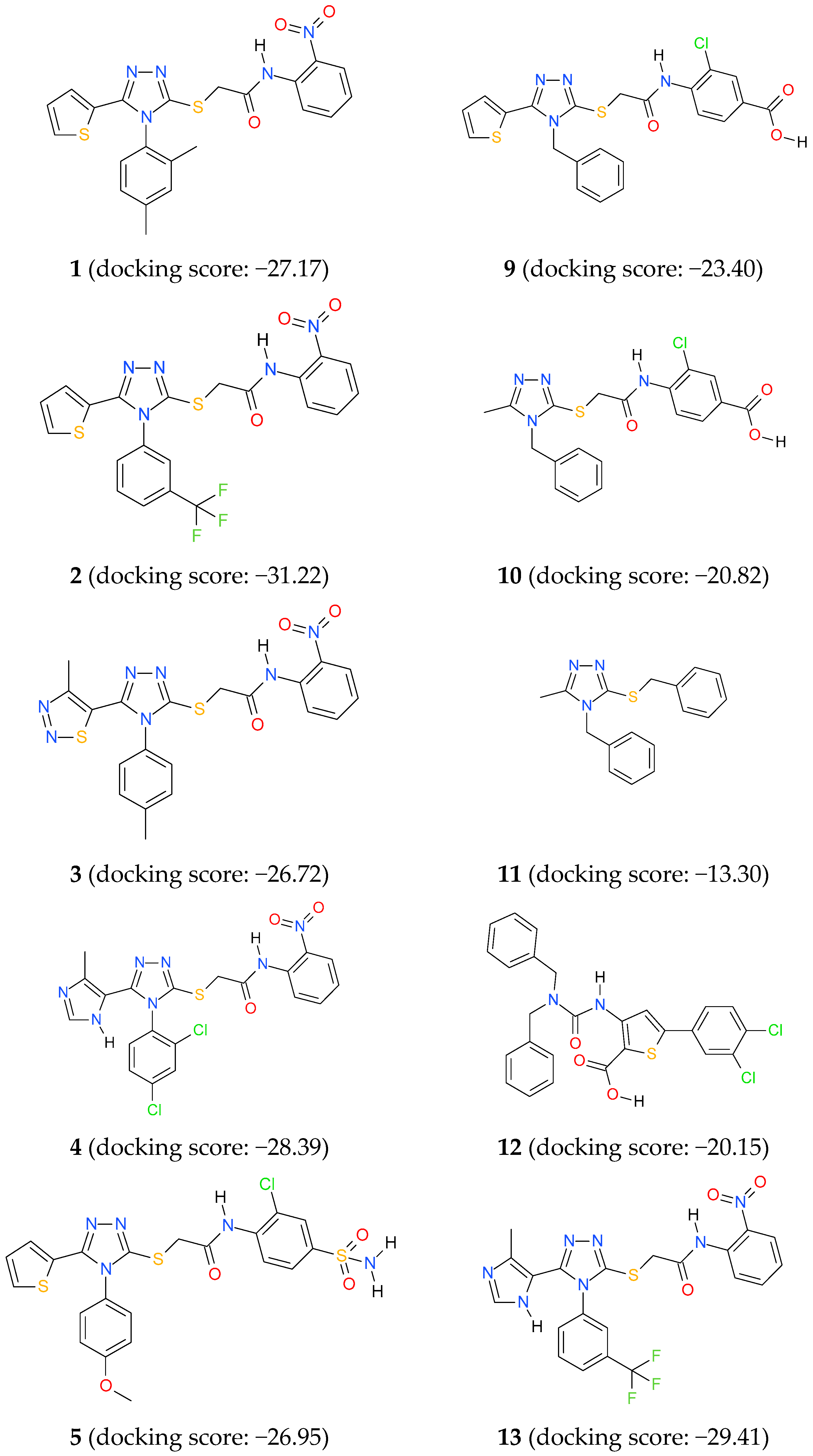
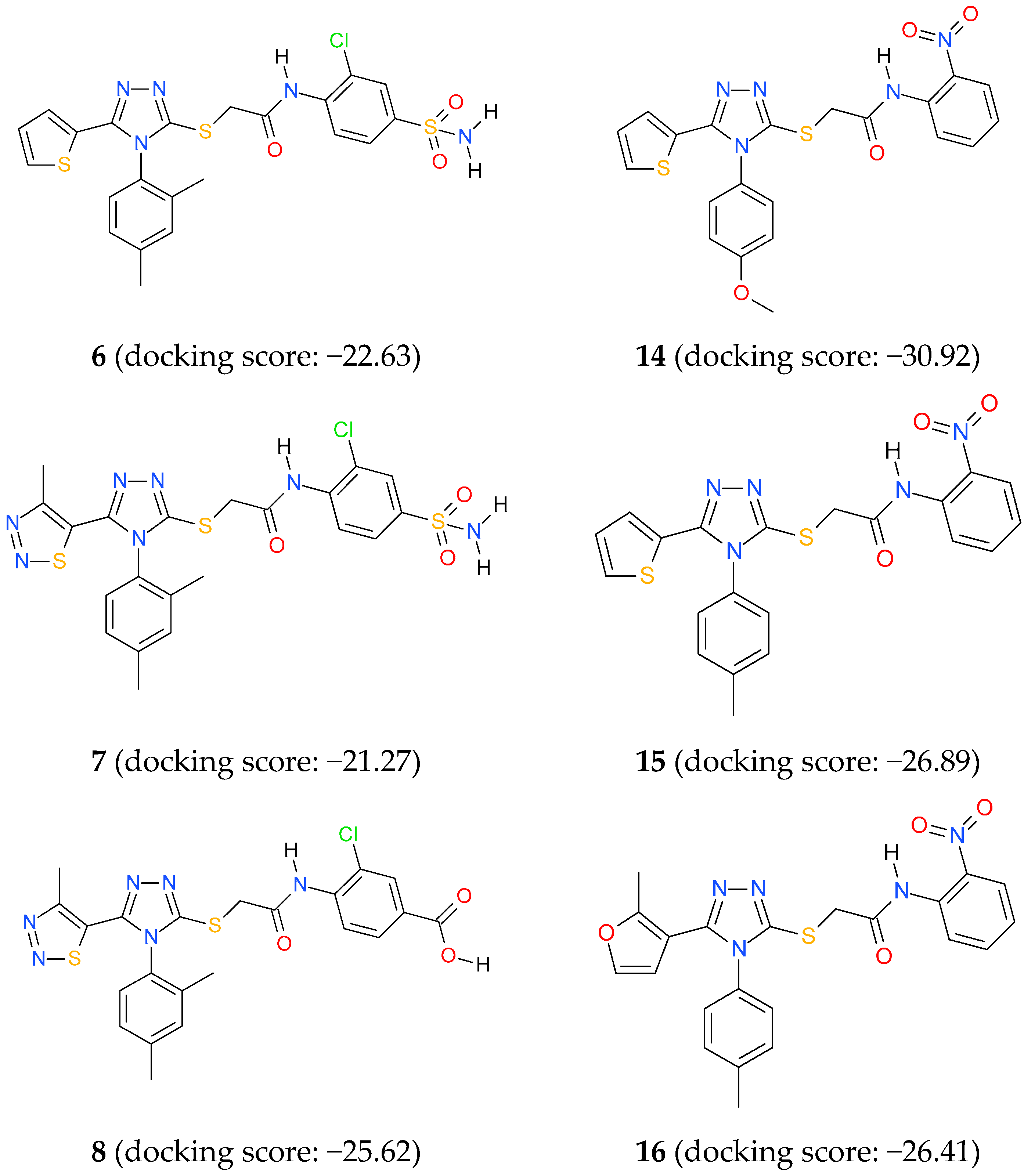
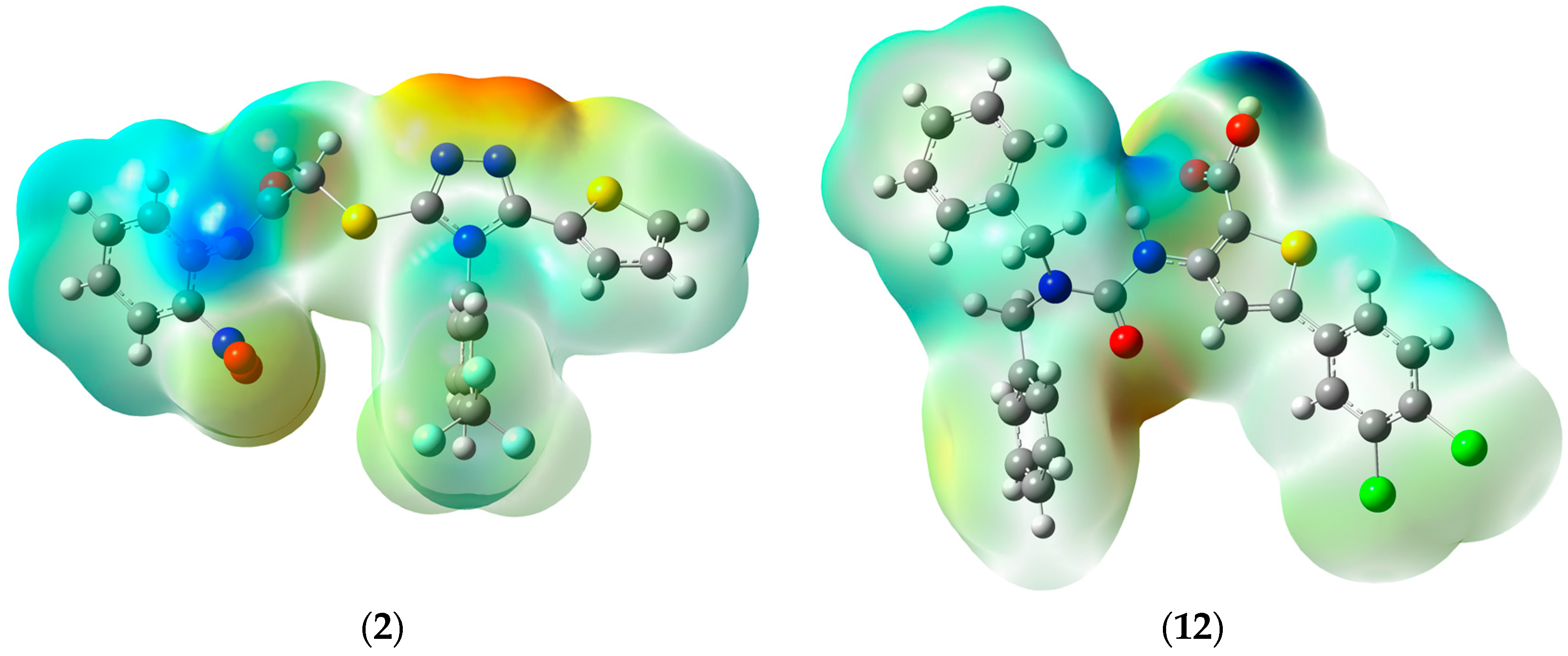
3.4. Docking Studies
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Février, M.; Dorgham, K.; Rebollo, A. CD4+ T cell depletion in human immunodeficiency virus (HIV) infection: Role of apoptosis. Viruses 2011, 3, 586–612. [Google Scholar] [CrossRef] [PubMed]
- Global Information and Education on HIV and AIDS. Available online: https://www.avert.org/global-hiv-and-aids-statistics (accessed on 20 September 2017).
- Buckheit, K.W.; Lu, Y.; Buckheit, R.W., Jr. Development of dual-acting pyrimidinediones as novel and highly potent topical anti-HIV microbicides. Antimicrob. Agents Chemother. 2011, 55, 5243–5254. [Google Scholar] [CrossRef] [PubMed]
- Berger, B.J.; Hussain, F.; Roistacher, K. Bacterial infections in HIV-infected patients. Infect. Dis. N. Am. 1994, 8, 449–465. [Google Scholar]
- Dejesus, E.; Young, B.; Morales-Ramirez, J.O.; Sloan, L.; Ward, D.J.; Flaherty, J.F.; Ebrahimi, R.; Maa, J.-F.; Reilly, K.; Ecker, J.; et al. Simplification of antiretroviral therapy toa single-tablet regimen consisting of Efavirenz, Emtricitabine, and Tenofovir Disoproxil Fumarate versus unmodified antiretroviral therapy in virologically suppressed HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 2009, 51, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Frączek, T.; Paneth, A.; Kamiński, K.; Krakowiak, A.; Paneth, P. Searching for novel scaffold of triazole non-nucleoside inhibitors ofHIV-1 reverse transcriptase. J. Enzym. Inhib. Med. Chem. 2016, 31, 481–489. [Google Scholar]
- Darst, S. New inhibitors targeting bacterial RNA polymerase. Trends Biochem. Sci. 2004, 29, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Chopra, I. Bacterial RNA polymerase: A promising target for the discovery of new antimicrobial agents. Curr. Opin. Investig. Drugs 2007, 8, 600–607. [Google Scholar] [PubMed]
- Mukhopadhyay, J.; Das, K.; Ismail, S.; Koppstein, D.; Jang, M.; Hudson, B.; Sarafianos, S.; Tuske, S.; Patel, J.; Jansen, R.; et al. The RNA polymerase “switch region” is a target for inhibitors. Cell 2008, 135, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Szilvay, A.M.; Stern, B.; Blichenberg, A.; Helland, D.E. Structural and functional similarities between HIV-1 reverse transcriptase and the Escherichia coli RNA polymerase β′ subunit. FEBS Lett. 2000, 484, 43–47. [Google Scholar] [CrossRef]
- Belogurov, G.A.; Vassylyeva, M.N.; Sevostyanova, A.; Appleman, J.R.; Xiang, A.X.; Lira, R.; Webber, S.E.; Klyuyev, S.; Nudler, E.; Artsimovitch, I.; et al. Transcription inactivation through local refolding of the RNA polymerase structure. Nature 2009, 457, 332–335. [Google Scholar] [CrossRef] [PubMed]
- Haebich, D.; von Nussbaum, F. Lost in transcription—Inhibition of RNA polymerase. Angew. Chem. Int. Ed. 2009, 48, 3397–3400. [Google Scholar] [CrossRef] [PubMed]
- Shea, R.O.; Moser, H.E. Physicochemical properties of antibacterial compounds: Implications for drug discovery. J. Med. Chem. 2008, 51, 2871–2878. [Google Scholar]
- Elgaher, W.A.M.; Sharma, K.K.; Haupenthal, J.; Saladini, F.; Pires, M.; Real, E.; Mely, Y.; Hartmann, R.W. Discovery and structure-based optimization of 2-ureidothiophene-3-carboxylic acids as dual bacterial RNA polymerase and viral reverse transcriptase inhibitors. J. Med. Chem. 2016, 59, 7212–7222. [Google Scholar] [CrossRef] [PubMed]
- Sahner, J.H.; Groh, M.; Negri, M.; Haupenthal, J.; Hartmann, R.W. Novel small molecule inhibitors targeting the “switch region” of bacterial RNAP: Structure-based optimization of a virtual screening hit. Eur. J. Med. Chem. 2013, 65, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Benno, R.; Hildegard, B. Splitting of hexamethylenetetramine addition compounds of N-haloacetylated substituted anilines be means of sulfur dioxide. Pharmazie 1949, 4, 149–150. [Google Scholar]
- ie, H.; Ng, D.; Savionv, S.N.; Dey, B.; Kwong, P.D.; Wyatt, R.; Smith, A.B., 3rd; Hendrickson, W.A. Structure-activity relationships inthe binding of chemically derivatized CD4 to gp120 from human immunodeficiency virus. J. Med. Chem. 2007, 50, 4898–4908. [Google Scholar]
- The European Committee on Antimicrobial Susceptibility Testing. European Committee for Antimicrobial Susceptibility Testing (EUCAST) Determination of Minimum Inhibitory Concentrations (MICs) of Antibacterial Agents by Broth Dilution; EUCAST discussion document E. Dis 5.1; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2003. [Google Scholar]
- Clinical and Laboratory Standards Institute. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; M27-S4; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Hyperchem; Version 8.0.3; HyperCube Inc.: Gainsville, FL, USA, 2007.
- Singh, U.C.; Kollman, P.A. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 1984, 5, 129–145. [Google Scholar] [CrossRef]
- Frisch, M.J. Gaussian 09; Revision A.02; Gaussian Inc.: Wallingford, UK, 2009. [Google Scholar]
Sample Availability: Samples of all compounds are available from the authors (A.P.). |

| 2 | 4 | 13 | 14 | 15 | 16 | Control * | |
|---|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | MIC; MBC (mg/mL) | ||||||
| S. aureus ATCC 6538 | 1; >1 | n.a. | 0.5; 2 | n.a. | 0.5; >1 | 1; >1 | 0.24; 0.24 |
| S. aureus ATCC 43300 | n.a. | 1; >1 | 0.25; 1 | n.a. | 1; >1 | 0.25; >1 | 0.24; 0.24 |
| S. aureus ATCC 25923 | n.a. | n.a. | 0.25; >1 | 0.5; >1 | 0.5; >1 | 0.5; >1 | 0.49; 0.49 |
| S. epidermidis ATCC 12228 | 1; >1 | 1; >1 | 0.06; 0.125 | 0.5; 2 | 0.003; 1 | 0.5; >1 | 0.49; 0.49 |
| B. subtilis ATCC 6633 | 1; >1 | 0.125; 1 | 0.125; 1 | 1; >1 | 1; >1 | 0.25; >1 | 0.03; 012 |
| B. cereus ATCC 10876 | 1; >1 | 1; >1 | 0.5; >1 | 1; >1 | 0.5; >1 | 1; >1 | 0.12; 0.12 |
| M. luteus ATCC 10240 | 1; 2 | 1; >1 | 0.125; 0.5 | 1; >1 | 1; >1 | 0.5; >1 | 0.98; 1.95 |
| Gram-negative bacteria | MIC; MBC (mg/mL) | ||||||
| E. coli ATCC 35218 | 1; >1 | n.a. | 1; >1 | 1; >1 | n.a. | 1; >1 | 0.015; 0.015 |
| E. coli ATCC 25922 | 1; >1 | n.a. | 1; >1 | 1; >1 | n.a. | 1; >1 | 0.008; 0.008 |
| S. typhimurium ATCC 14028 | 1; >1 | n.a. | 1; >1 | 1; >1 | n.a. | 1; >1 | 0.008; 0.008 |
| K. pneumoniae ATCC 13883 | n.a. | n.a. | 1; >1 | n.a. | n.a. | 1; >1 | 0.015; 0.015 |
| P. mirabilis ATCC 12453 | n.a. | n.a. | 1; >1 | n.a. | n.a. | n.a. | 0.015; 0.015 |
| P. aeruginosa ATCC 9027 | 1; >1 | 1; >1 | 1; >1 | 1; >1 | 1; >1 | 1; >1 | 0.12; 0.24 |
| B. bronchiseptica ATCC 4617 | 1; >1 | 0.5; >1 | 0.5; >1 | 0.5; >1 | 0.5; >1 | 0.5; >1 | 0.12; 0.12 |
| Yeasts | MIC; MBC (mg/mL) | ||||||
| C. parapsilosis ATCC 22019 | 0.5; 1 | 0.5; 1 | 0.5; 1 | 0.5; 1 | 0.5; 1 | 0.25; 1 | 1.95; 1.95 |
| C. albicans ATCC 2091 | 0.5; 1 | 0.5; 0.5 | 0.5; 0.5 | 0.5; 1 | 0.5; 0.5 | 0.125; 0.25 | 0.98; 0.98 |
| C. albicans ATCC 10231 | 1; 1 | 1; 1 | 0.5; 1 | 1; 1 | 0.5; 1 | 1; 1 | 0.98; 1.95 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paneth, A.; Frączek, T.; Grzegorczyk, A.; Janowska, D.; Malm, A.; Paneth, P. A Search for Dual Action HIV-1 Reverse Transcriptase, Bacterial RNA Polymerase Inhibitors. Molecules 2017, 22, 1808. https://doi.org/10.3390/molecules22111808
Paneth A, Frączek T, Grzegorczyk A, Janowska D, Malm A, Paneth P. A Search for Dual Action HIV-1 Reverse Transcriptase, Bacterial RNA Polymerase Inhibitors. Molecules. 2017; 22(11):1808. https://doi.org/10.3390/molecules22111808
Chicago/Turabian StylePaneth, Agata, Tomasz Frączek, Agnieszka Grzegorczyk, Dominika Janowska, Anna Malm, and Piotr Paneth. 2017. "A Search for Dual Action HIV-1 Reverse Transcriptase, Bacterial RNA Polymerase Inhibitors" Molecules 22, no. 11: 1808. https://doi.org/10.3390/molecules22111808





