Phenolic Compounds from Belamcanda chinensis Seeds
Abstract
:1. Introduction
2. Results and Discussion
2.1. Structure Elucidation of the Compounds
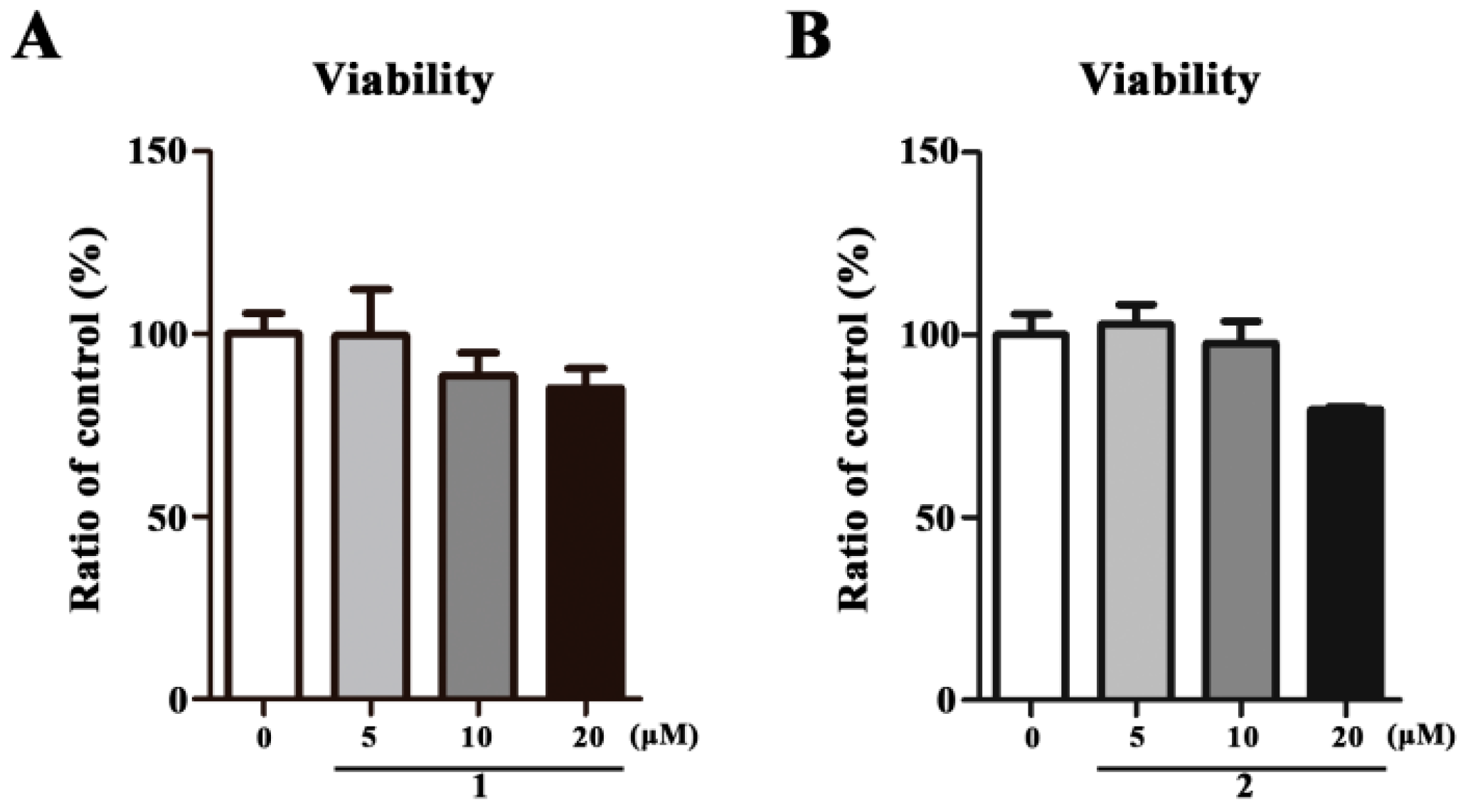
2.2. Cell Viability Assay
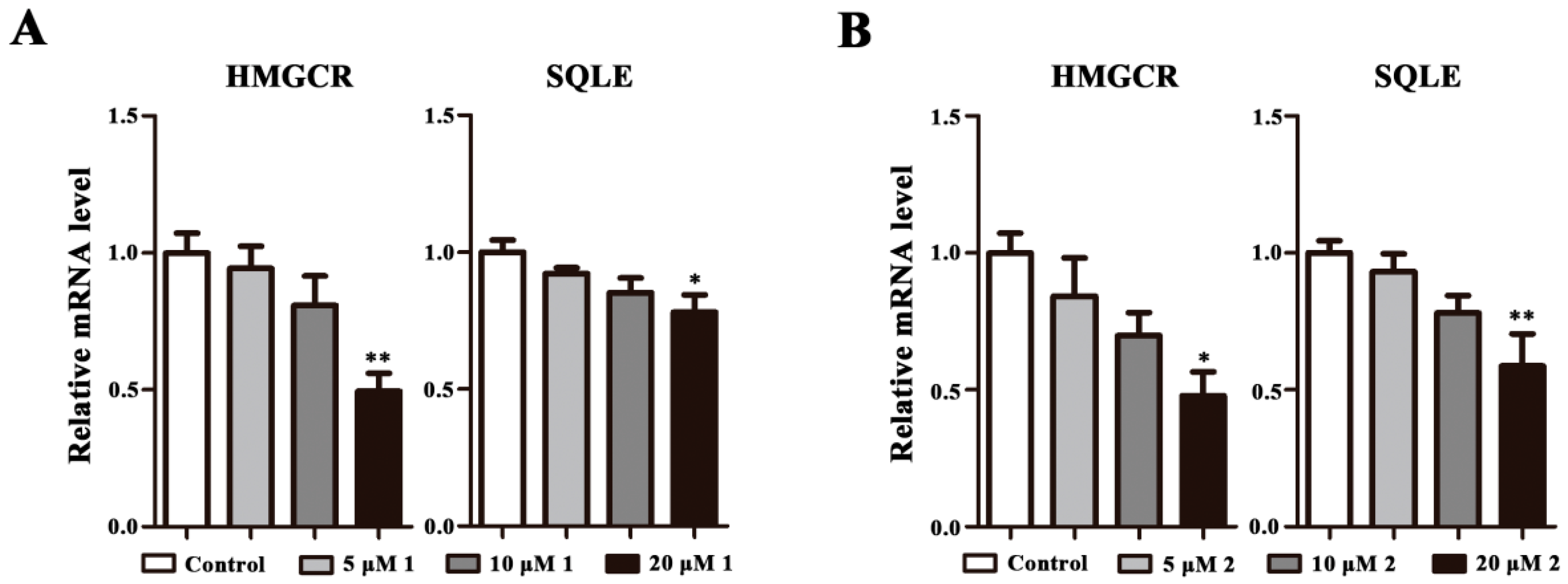
2.3. Effects on HMGCR and SQLE mRNA Expression
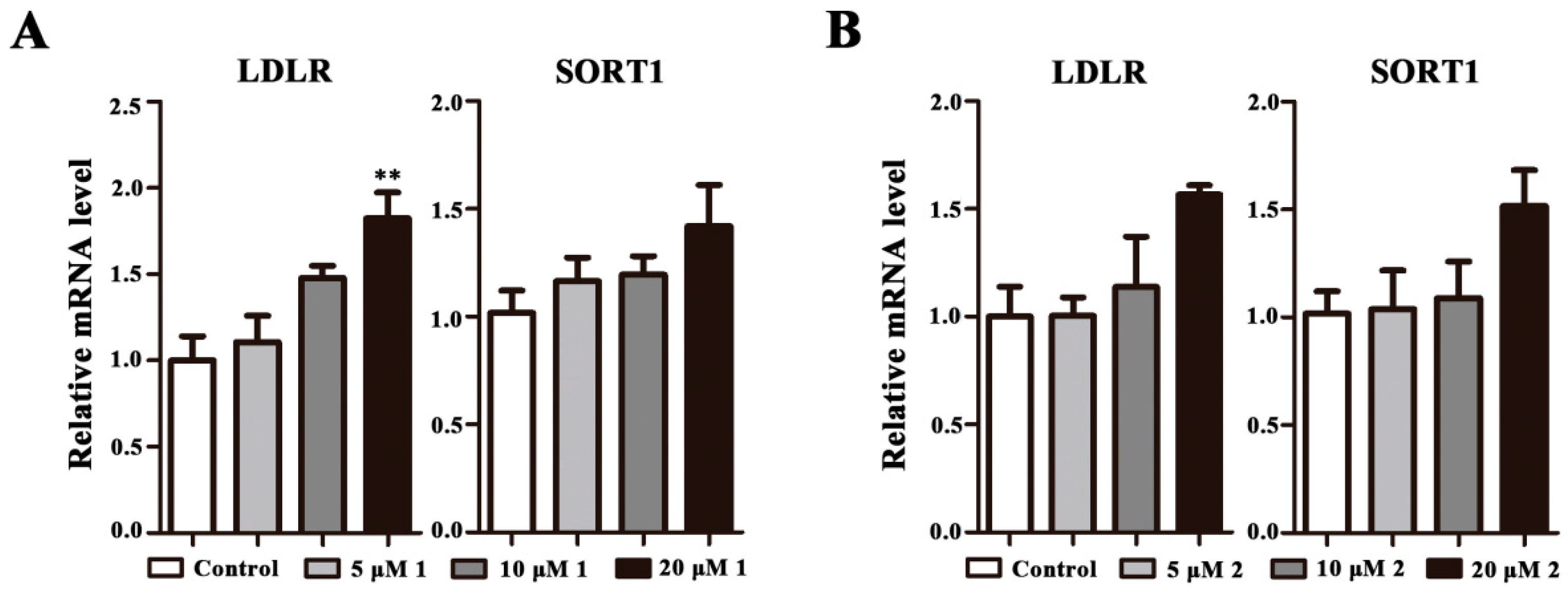
2.4. Effects on LDLR and SORT1 mRNA Expression
2.5. Discussion
3. Experimental Section
3.1. General Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Compound Characterization Data
3.5. Acid Hydrolysis and Sugar Analysis
3.6. Cell Culture
3.7. Cell Viability Assay
3.8. RNA Isolation and qPCR
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chinese Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; Chinese Medical Science and Technology Press: Beijing, China, 2010; Volume 1, pp. 267–268. [Google Scholar]
- Monthakantirat, O.; De Eknamkul, W.; Umehara, K.; Yoshinaga, Y.; Miyase, T.; Warashina, T.; Noguchi, H. Phenolic Constituents of the Rhizomes of the Thai Medicinal Plant Belamcanda chinensis with Proliferative Activity for Two Breast Cancer Cell Lines. J. Nat. Prod. 2005, 68, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Fukuyama, Y.; Kiriyama, Y.; Okino, J.; Kodama, M. Belamcandaquinones A and B, novel dimeric 1,4-benzoquinone derivatives possessing cyclooxygenase inhibitory activity. Tetrahedron Lett. 1993, 34, 7633–7636. [Google Scholar] [CrossRef]
- Ito, H.; Onoue, S.; Miyake, Y.; Yoshida, T. Iridal-type triterpenoids with ichthyotoxic activity from Belamcanda chinensis. J. Nat. Prod. 1999, 62, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.; Jin, L. Advances in research on phytochemical constituents and pharmacology of Belamcanda chinensis. Pharm. Care Res. 2008, 8, 358–361. [Google Scholar]
- Ai, J.; Nie, J.; He, J.; Guo, Q.; Li, M.; Lei, Y.; Liu, Y.H.; Zhou, Z.H.; Zhu, F.X.; Liang, M.; et al. GQ5 Hinders Renal Fibrosis in Obstructive Nephropathy by Selectively Inhibiting TGF-β–Induced Smad3 Phosphorylation. J. Am. Soc. Nephrol. 2015, 26, 1827–1838. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.Y.; Ai, J.; Wang, X.L.; Qiu, F.G.; Lv, Q.; Fang, P.; Hou, F.F.; Yan, Y.M.; Cheng, Y.X. Isolation of lingzhifuran A and lingzhilactones D–F from Ganoderma lucidum as specific Smad3 phosphorylation inhibitors and total synthesis of lingzhifuran A. RSC Adv. 2016, 6, 77887–77897. [Google Scholar] [CrossRef]
- Seki, K.; Haga, K.; Kaneko, R. Belamcandones AD, dioxotetrahydrodibenzofurans from Belamcanda chinensis. Phytochemistry 1995, 38, 703–709. [Google Scholar] [CrossRef]
- Agrawal, P.K. NMR-spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 1992, 31, 3307–3330. [Google Scholar] [CrossRef]
- Zhang, L.; Liao, C.C.; Huang, H.C.; Shen, Y.C.; Yang, L.M.; Kuo, Y.H. Antioxidant phenylpropanoid glycosides from Smilax bracteata. Phytochemistry 2008, 69, 1398–1404. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.L.; Zhang, L.J.; Chen, R.Y.; Kuo, L.M.Y.; Huang, J.P.; Huang, H.C.; Lee, K.H.; Wu, Y.C.; Kuo, Y.H. Antioxidant and Anti-inflammatory Phenylpropanoid Derivatives from Calamus quiquesetinervius. J. Nat. Prod. 2010, 73, 1482–1488. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kim, C.S.; Park, Y.J.; Moon, E.; Choi, S.U.; Lee, J.H.; Kim, S.Y.; Lee, K.R. Anti-inflammatory and antitumor phenylpropanoid sucrosides from the seeds of Raphanus sativus. Bioorg. Med. Chem. Lett. 2015, 25, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.R.; Lee, S.; Eom, H.J.; Yu, J.S.; Lee, S.R. Anti-inflammatory and antitumor phenylpropanoid sucrosides from the seeds of Raphanus sativus. Planta Med. 2015, 81, 932. [Google Scholar] [CrossRef]
- Rohaiza, S.; Yaacob, W.A.; Din, L.B.; Nazlina, I. Cytotoxic oligostilbenes from Shorea hopeifolia. Afr. J. Pharm. Pharmacol. 2011, 5, 1272–1277. [Google Scholar] [CrossRef]
- Cai, X.F.; Lee, I.S.; Dat, N.T.; Shen, G.; Kang, J.S.; Kim, D.H.; Kim, Y.H. Inhibitory lignans against NFAT transcription factor from acanthopanax koreanum. Arch. Pharm. Res. 2004, 27, 738–741. [Google Scholar] [CrossRef] [PubMed]
- Miyazawa, M.; Hisama, M. Antimutagenic activity of flavonoids from Chrysanthemum morifolium. Biosci. Biotechnol. Biochem. 2003, 67, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.J.; Wang, J.; Chen, S.; Luo, S.; Xu, X.M. Study on the Chemical Constituents of Iris tectorum Maxim. Nat. Prod. Res. Dev. 2008, 20, 444–446. [Google Scholar]
- Ito, H.; Onoue, S.; Yoshida, T. Isoflavonoids from Belamcanda chinensis. Chem. Pharm. Bull. 2001, 49, 1229–1231. [Google Scholar] [CrossRef] [PubMed]
- Howe, V.; Sharpe, L.J.; Prabhu, A.V.; Brown, A.J. New insights into cellular cholesterol acquisition: Promoter analysis of human HMGCR and SQLE, two key control enzymes in cholesterol synthesis. BBA. 2017, 1862, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, Z.Q.; Jiang, S.; Hu, W.; Li, T.; Di, S.Y.; Wang, D.J.; Yang, Y. A global perspective on FOXO1 in lipid metabolism and lipid-related diseases. Prog. Lipid Res. 2017, 66, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Murase, T.; Yokoi, Y.; Misawa, K.; Ominami, H.; Suzuki, Y.; Shibuya, Y.; Hase, T. Coffee polyphenols modulate whole-body substrate oxidation and suppress postprandial hyperglycaemia, hyperinsulinaemia and hyperlipidaemia. Br. J. Nutr. 2012, 107, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Priore, P.; Cavallo, A.; Gnoni, A.; Damiano, F.; Gnoni, G.V.; Siculella, L. Modulation of Hepatic Lipid Metabolism by Olive Oil and its Phenols in Nonalcoholic Fatty Liver Disease. Iubmb Life 2015, 67, 1–9. [Google Scholar]
- Priore, P.; Siculella, L.; Gnoni, G.V. Extra virgin olive oil phenols down-regulate lipid synthesis in primary-cultured rat-hepatocytes. J. Nutr. Biochem. 2014, 25, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.F. Single nucleotide resolution of sterol regulatory region in promoterfor 3-hydroxy-3-methylglutaryl coenzyme A reductase. J. Biol. Chem. 1991, 266, 13947–13951. [Google Scholar]
- Gill, S.; Stevenson, J.; Kristiana, I.; Brown, A.J. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011, 13, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Puleston, D.J.; Villa, M.; Pearce, E.L. Ancillary Activity: Beyond Core Metabolism in Immune Cells. Cell Metab. 2017, 26, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.S.; Goldstein, J.L. A receptor-mediated pathway for cholesterol homeostasis. Science 1986, 232, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Bi, L.; Chiang, J.Y.; Ding, W.X.; Dunn, W.; Roberts, B.; Li, T.G. Saturated fatty acids activate ERK signaling to downregulate hepatic sortilin 1 in obese and diabetic mice. J. Lipid Res. 2013, 54, 2754–2762. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 3–7 are available from the authors. |


| 1 | 2 | ||||
|---|---|---|---|---|---|
| Position | δH (J/Hz) | δC Mult | Position | δH (J/Hz) | δC Mult |
| 1 | 4.37, s | 66.7, t | 1 | 4.39, s | 66.5, t |
| 2 | 103.5, s | 2 | 103.5, s | ||
| 3 | 5.57, d, (8.3) | 79.9, d | 3 | 5.62, d, (8.3) | 79.9, d |
| 4 | 4.46, t, (8.3) | 73.3, d | 4 | 4.44, t, (8.3) | 73.5, d |
| 5 | 3.98, m | 84.2, d | 5 | 4.00, m | 84.3, d |
| 6 | 3.85, m | 63.0, t | 6 | 3.86, m | 63.1, t |
| 1’ | 5.60, d, (3.6) | 93.3, s | 1’ | 5.66, d, (3.5) | 92.2, s |
| 2’ | 3.55, dd, (9.8, 3.6) | 73.1, d | 2’ | 3.74, m | 71.1, d |
| 3’ | 3.89, m | 72.8, d | 3’ | 5.47, t, (9.8) | 74.5, d |
| 4’ | 4.93, m | 72.7, d | 4’ | 5.12, t, (9.8) | 70.2, d |
| 5’ | 4.21, m | 72.6, d | 5’ | 4.30, brd (4.3) | 72.4, d |
| 6′ | 3.60, m | 62.4, t | 6′ OAc-3’ | 3.72, m | 61.9, t |
| 3.71, brd, (12.1) | 1.98, s | 172.3, s | |||
| 20.9, q | |||||
| 1’’ | 127.5, s | 1’’ | 127.6, s | ||
| 2’’ | 7.15, d, (1.6) | 111.6, d | 2’’ | 7.11, d, (1.5) | 111.7, d |
| 3’’ | 149.3, s | 3’’ | 149.2, s | ||
| 4’’ | 150.7, s | 4’’ | 150.8, s | ||
| 5’’ | 6.75, d, (8.2) | 116.4, d | 5’’ | 6.72, d, (8.2) | 116.3, d |
| 6’’ | 7.10, dd, (8.2, 1.6) | 124.5, d | 6’’ | 7.03, dd, (8.2, 1.5) | 124.4, d |
| 7’’ | 7.76, d, (15.9) | 114.8, d | 7’’ | 7.65, d, (15.9) | 114.9, d |
| 8’’ | 6.46, d, (15.9) | 148.0, d | 8’’ | 6.35, d, (15.9) | 147.7, d |
| 9’’ | 168.5, s | 9’’ | 168.5, s | ||
| OCH3 | 3.80, s | 56.4, s | OCH3 | 3.80, s | 56.3, s |
| 1’’’ | 127.5, s | 1’’’ | 127.6, s | ||
| 2’’’ | 7.05, d, (1.6) | 111.7, d | 2’’’ | 7.03, d, (1.5) | 111.6, d |
| 3’’’ | 149.3, s | 3’’’ | 149.3, s | ||
| 4’’’ | 150.7, s | 4’’’ | 150.6, s | ||
| 5’’’ | 6.80, d, (8.2) | 116.6, d | 5’’’ | 6.70, d, (8.2) | 116.4, d |
| 6’’’ | 6.87, dd, (8.2, 1.6) | 124.0, d | 6’’’ | 7.11, dd, (8.2, 1.5) | 124.6, d |
| 7’’’ | 7.57, d, (15.9) | 115.0, d | 7’’’ | 7.74, d, (15.9) | 115.0, d |
| 8’’’ | 6.19, d, (15.9) | 147.5, d | 8’’’ | 6.45, d, (15.9) | 148.1, d |
| 9’’’ | 168.2, s | 9’’’ | 168.2, s | ||
| OCH3 | 3.83, s | 56.4, s | OCH3 | 3.76, s | 56.4, s |
| 1’’’’ | 127.6, s | 1’’’’ | 127.2, s | ||
| 2’’’’ | 7.15, d, (1.6) | 111.7, d | 2’’’’ | 7.15, d, (1.5) | 111.8, d |
| 3’’’’ | 149.4, s | 3’’’’ | 149.2, s | ||
| 4’’’’ | 151.0, s | 4’’’’ | 150.9, s | ||
| 5’’’’ | 6.78, d, (8.2) | 116.6, d | 5’’’’ | 6.73, d, (8.2) | 116.6, d |
| 6’’’’ | 7.06, dd, (8.2, 1.6) | 124.0, d | 6’’’’ | 7.03, dd, (8.2, 1.5) | 124.3, d |
| 7’’’’ | 7.67, d, (15.9) | 114.9, d | 7’’’’ | 7.53, d, (15.9) | 114.2 , d |
| 8’’’’ | 6.38, d, (15.9) | 147.6, d | 8’’’’ | 6.12, d, (15.9) | 148.0, d |
| 9’’’’ | 168.5, s | 9’’’’ | 167.8, s | ||
| OCH3 | 3.84, s | 56.4, s | OCH3 | 3.78, s | 56.4, s |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, Y.-Y.; Liu, Y.; Yan, Y.-M.; Lu, X.-F.; Cheng, Y.-X. Phenolic Compounds from Belamcanda chinensis Seeds. Molecules 2018, 23, 580. https://doi.org/10.3390/molecules23030580
Song Y-Y, Liu Y, Yan Y-M, Lu X-F, Cheng Y-X. Phenolic Compounds from Belamcanda chinensis Seeds. Molecules. 2018; 23(3):580. https://doi.org/10.3390/molecules23030580
Chicago/Turabian StyleSong, Ying-Ying, Ying Liu, Yong-Ming Yan, Xi-Feng Lu, and Yong-Xian Cheng. 2018. "Phenolic Compounds from Belamcanda chinensis Seeds" Molecules 23, no. 3: 580. https://doi.org/10.3390/molecules23030580




