14-3-3: A Case Study in PPI Modulation
Abstract
:1. Introduction
2. 14-3-3/Ligand Interactions
3. 14-3-3 PPIs in Human Diseases
4. Stabilizers of 14-3-3 PPIs
4.1. Natural Products
4.1.1. Fusicoccin-A
4.1.2. Cotylenin-A
4.1.3. Mizoribine (or Bredinin)
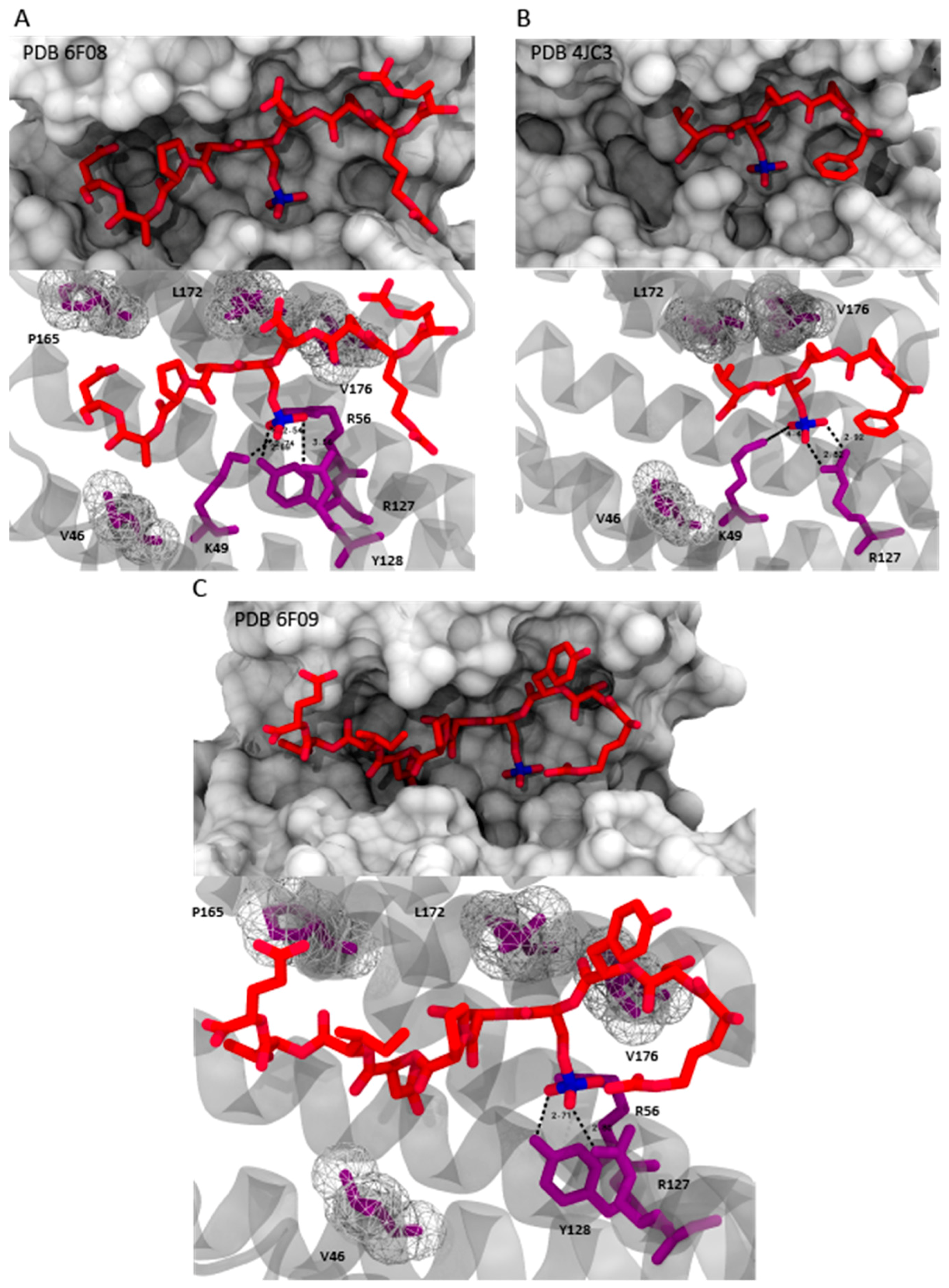
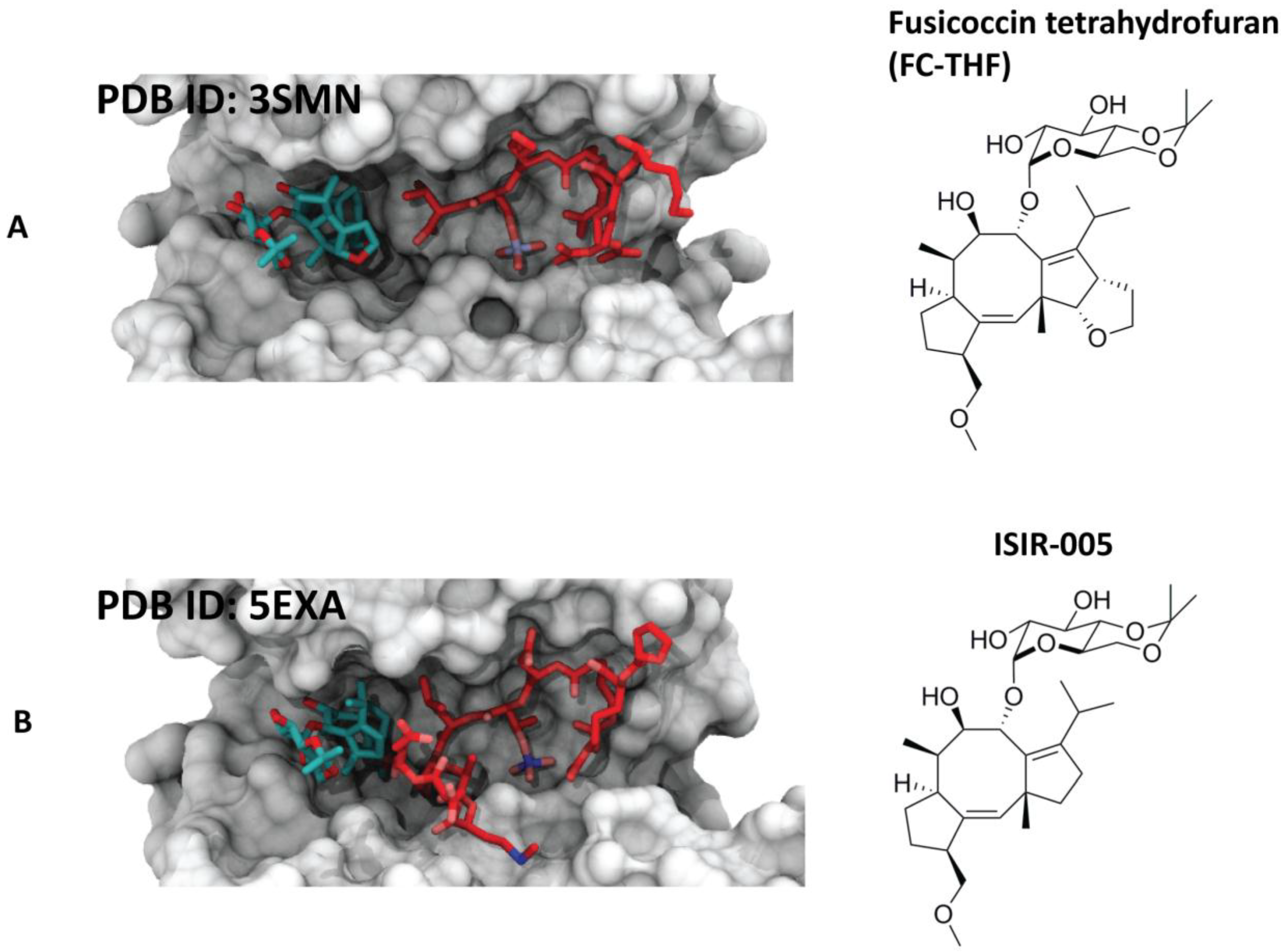
4.2. Semisynthetic Fucicoccanes
4.3. Synthesis Products
4.3.1. Pyrrolidone1 and Pyrazole 37
4.3.2. Adenosine Monophosphate (AMP)
4.3.3. The Molecular Tweezer CLR01
5. Inhibitors of 14-3-3 PPIs
6. Conclusions
Funding
Conflicts of Interest
References
- Moore, B.W.; Perez, V.J.; Carlson, F.D. (Eds.) Physiological and Biochemical Aspects of Nervous Integration; Prentice-Hall Inc.; The Marine Biological Laboratory: Woods Hole, MA, USA, 1967; pp. 343–359. [Google Scholar]
- Aitken, A. 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 2006, 16, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, T.; Isobe, T.; Okuyama, T.; Takahashi, N.; Araki, K.; Kuwano, R.; Takahashi, Y. Molecular cloning of cDNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc. Natl. Acad. Sci. USA 1988, 85, 7084–7088. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shakes, D.C. Molecular evolution of the 14-3-3 protein family. J. Mol. Evol. 1996, 43, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Cau, Y.; Valensin, D.; Mori, M.; Draghi, S.; Botta, M. Structure, function, involvement in diseases and targeting of 14-3-3 proteins: An update. Curr. Med. Chem. 2017, 25, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Stevers, L.M.; Sijbesma, E.; Botta, M.; MacKintosh, C.; Obsil, T.; Landrieu, I.; Cau, Y.; Wilson, A.J.; Karawajczyk, A.; Eickhoff, J.; et al. Modulators of 14-3-3 Protein–Protein Interactions. J. Med. Chem. 2017, 61, 3755–3778. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Bienkowska, J.; Petosa, C.; Collier, R.J.; Fu, H.; Liddington, R. Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 1995, 376, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.H.; Ley, S.; Aitken, A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: Implications for function as adapter proteins. FEBS Lett. 1995, 368, 55–58. [Google Scholar] [CrossRef]
- Bridges, D.; Moorhead, G.B.G. 14-3-3 proteins: A number of functions for a numbered protein. Sci. STKE Signal Transduct. Knowl. Environ. 2005, 2005, re10. [Google Scholar] [CrossRef] [PubMed]
- Bartel, M.; Schäfer, A.; Stevers, L.M.; Ottmann, C. Small molecules, peptides and natural products: Getting a grip on 14-3-3 protein–protein modulation. Future Med. Chem. 2014, 6, 903–921. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.; Bartel, M.; Sies, K.; Halbach, S.; Higuchi, Y.; Haranosono, Y.; Brummer, T.; Kato, N.; Ottmann, C. Small-Molecule Stabilization of the 14-3-3/Gab2 Protein–Protein Interaction (PPI) Interface. ChemMedChem 2016, 11, 911–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, T.; Fournier, A.E.; Yamagata, K. Neuroprotective function of 14-3-3 proteins in neurodegeneration. BioMed Res. Int. 2013, 564534. [Google Scholar] [CrossRef] [PubMed]
- Yaffe, M.B.; Rittinger, K.; Volinia, S.; Caron, P.R.; Aitken, A.; Leffers, H.; Gamblin, S.J.; Smerdon, S.J.; Cantley, L.C. The Structural Basis for 14-3-3: Phosphopeptide Binding Specificity. Cell 1997, 91, 961–971. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Liddington, R.; Fu, H. Mutations in the hydrophobic surface of an amphipathic groove of 14-3-3zeta disrupt its interaction with Raf-1 kinase. J. Biol. Chem. 1998, 273, 16297–16304. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Smerdon, S.J.; Jones, D.H.; Dodson, G.G.; Soneji, Y.; Aitken, A.; Gamblin, S.J. Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 1995, 376, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Coblitz, B.; Wu, M.; Shikano, S.; Li, M. C-terminal binding: An expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006, 580, 1531–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, M.P.; Geraghty, K.M.; Wong, B.H.C.; Wood, N.T.; Campbell, D.G.; Morrice, N.; Mackintosh, C. 14-3-3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem. J. 2004, 379, 395–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardino, A.K.; Smerdon, S.J.; Yaffe, M.B. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: A comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin. Cancer Biol. 2006, 16, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Ottmann, C.; Weyand, M.; Sassa, T.; Inoue, T.; Kato, N.; Wittinghofer, A.; Oecking, C. A structural rationale for selective stabilization of anti-tumor interactions of 14-3-3 proteins by cotylenin A. J. Mol. Biol. 2009, 386, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Ballone, A.; Centorrino, F.; Wolter, M.; Ottmann, C. Structural characterization of 14-3-3ζ in complex with the human Son of sevenless homolog 1 (SOS1). J. Struct. Biol. 2018. [Google Scholar] [CrossRef] [PubMed]
- De Vries-van Leeuwen, I.J.; da Costa Pereira, D.; Flach, K.D.; Piersma, S.R.; Haase, C.; Bier, D.; Yalcin, Z.; Michalides, R.; Feenstra, K.A.; Jiménez, C.R.; et al. Interaction of 14-3-3 proteins with the estrogen receptor alpha F domain provides a drug target interface. Proc. Natl. Acad. Sci. USA 2013, 110, 8894–8899. [Google Scholar] [CrossRef] [PubMed]
- Centorrino, F.; Ballone, A.; Wolter, M.; Ottmann, C. Biophysical and structural insight into the USP8/14-3-3 interaction. FEBS Lett. 2018, 592, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Synowsky, S.; Tinti, M.; MacKintosh, C. The capture of phosphoproteins by 14-3-3 proteins mediates actions of insulin. Trends Endocrinol. Metab. 2011, 22, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The 14-3-3 cancer connection. Nat. Rev. Cancer 2003, 3, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabási, A.-L. Interactome Networks and Human Disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Nooren, I.M.A.; Thornton, J.M. Diversity of protein-protein interactions. EMBO J. 2003, 22, 3486–3492. [Google Scholar] [CrossRef] [PubMed]
- Aghazadeh, Y.; Papadopoulos, V. The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov. Today 2016, 21, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Wilker, E.; Yaffe, M.B. 14-3-3 Proteins—A focus on cancer and human disease. J. Mol. Cell. Cardiol. 2004, 37, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Steinacker, P.; Aitken, A.; Otto, M. 14-3-3 proteins in neurodegeneration. Semin. Cell Dev. Biol. 2011, 22, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Ottmann, C.; Fournier, A.E. 14-3-3 adaptor protein-protein interactions as therapeutic targets for CNS diseases. Pharmacol. Res. 2017, 125, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, M.; Sobue, K.; Paudel, H.K. 14-3-3zeta is an effector of tau protein phosphorylation. J. Biol. Chem. 2000, 275, 25247–25254. [Google Scholar] [CrossRef] [PubMed]
- Sadik, G.; Tanaka, T.; Kato, K.; Yamamori, H.; Nessa, B.N.; Morihara, T.; Takeda, M. Phosphorylation of tau at Ser214 mediates its interaction with 14-3-3 protein: Implications for the mechanism of tau aggregation. J. Neurochem. 2009, 108, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Seit-Nebi, A.S.; Gusev, N.B. Effect of phosphorylation on interaction of human tau protein with 14-3-3zeta. Biochem. Biophys. Res. Commun. 2009, 379, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Gusev, N.B. Probable participation of 14-3-3 in tau protein oligomerization and aggregation. J. Alzheimers Dis. JAD 2011, 27, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Stevers, L.M.; Lam, C.V.; Leysen, S.F.R.; Meijer, F.A.; van Scheppingen, D.S.; de Vries, R.M.J.M.; Carlile, G.W.; Milroy, L.G.; Thomas, D.Y.; Brunsveld, L.; et al. Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR. Proc. Natl. Acad. Sci. USA 2016, 113, E1152–E1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reincke, M.; Sbiera, S.; Hayakawa, A.; Theodoropoulou, M.; Osswald, A.; Beuschlein, F.; Komada, M. Mutations in the deubiquitinase gene USP8 cause Cushing’s disease. Nat. Gen. 2015, 47, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhu, W.; Huang, J.; Wang, X.; Sun, X.; Zhan, B.; Zhu, X. Partially protective immunity induced by the 14-3-3 protein from Trichinella spiralis. Vet. Parasitol. 2016, 231, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Giordanetto, F.; Schäfer, A.; Ottmann, C. Stabilization of protein–protein interactions by small molecules. Drug Discov. Today 2014, 19, 1812–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, C.; Higuchi, Y.; Koschinsky, K.; Bartel, M.; Schumacher, B.; Thiel, P.; Nitta, H.; Preisig-Müller, R.; Schlichthörl, G.; Renigunta, V.; et al. A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface. Chem. Biol. 2013, 20, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Oecking, C.; Eckerskorn, C.; Weiler, E.W. The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 1994, 352, 163–166. [Google Scholar] [CrossRef] [Green Version]
- Würtele, M.; Jelich-Ottmann, C.; Wittinghofer, A.; Oecking, C. Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 2003, 22, 987–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molzan, M.; Schumacher, B.; Ottmann, C.; Baljuls, A.; Polzien, L.; Weyand, M.; Thiel, P.; Rose, R.; Rose, M.; Kuhenne, P.; et al. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol. Cell. Biol. 2010, 30, 4698–4711. [Google Scholar] [CrossRef] [PubMed]
- Molzan, M.; Kasper, S.; Röglin, L.; Skwarczynska, M.; Sassa, T.; Inoue, T.; Breitenbuecher, F.; Ohkanda, J.; Kato, N.; Schuler, M.; et al. Stabilization of Physical RAF/14-3-3 Interaction by Cotylenin A as Treatment Strategy for RAS Mutant Cancers. ACS Chem. Biol. 2013, 8, 1869–1875. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Higuchi, Y.; Yoneyama, T.; Lin, B.; Nunomura, K.; Honma, Y.; Kato, N. Semisynthesis and biological evaluation of a cotylenin A mimic derived from fusicoccin A. Bioorg. Med. Chem. Lett. 2018, 28, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Kamata, K.; Okubo, M.; Ishigamori, E.; Masaki, Y.; Uchida, H.; Watanabe, K.; Kashiwagi, N. Immunosuppressive effect of bredinin on cell-mediated and humoral immune reactions in experimental animals. Transplantation 1983, 35, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Wakui, H.; Gustafsson, J.A.; Zilliacus, J.; Itoh, H. Functional interaction of the immunosuppressant mizoribine with the 14-3-3 protein. Biochem. Biophys. Res. Commun. 2000, 274, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.; Erdmann, S.; Bovens, S.; Wolf, A.; Rose, M.; Hennig, S.; Waldmann, H.; Ottmann, C. Identification and Structure of Small-Molecule Stabilizers of 14-3-3 Protein–Protein Interactions. Angew. Chem. Int. Ed. 2010, 49, 4129–4132. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.; Rose, R.; Hedberg, C.; Waldmann, H.; Ottmann, C. An Optimised Small-Molecule Stabiliser of the 14-3-3–PMA2 Protein–Protein Interaction. Chem. Eur. J. 2012, 18, 6520–6527. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Jung, H.; Nakagawa, T.; Pawlosky, R.; Takeshima, T.; Lee, W.-R.; Sakiyama, H.; Laxman, S.; Wynn, R.M.; Tu, B.P.; et al. Metabolite Regulation of Nuclear Localization of Carbohydrate-response Element-binding Protein (ChREBP). Role of AMP as an Allosteric Inhibitor. J. Biol. Chem. 2016, 291, 10515–10527. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.; Rose, R.; Bravo-Rodriguez, K.; Bartel, M.; Ramirez-Anguita, J.M.; Dutt, S.; Wilch, C.; Klärner, F.-G.; Sanchez-Garcia, E.; Schrader, T.; et al. Molecular tweezers modulate 14-3-3 protein-protein interactions. Nat. Chem. 2013, 5, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Bier, D.; Mittal, S.; Bravo-Rodriguez, K.; Sowislok, A.; Guillory, X.; Briels, J.; Heid, C.; Bartel, M.; Wettig, B.; Brunsveld, L.; et al. The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface. J. Am. Chem. Soc. 2017, 139, 16256–16263. [Google Scholar] [CrossRef] [PubMed]
- Galaktionov, K.; Beach, D. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: Evidence for multiple roles of mitotic cyclins. Cell 1991, 67, 1181–1194. [Google Scholar] [CrossRef]
- Lopez-Girona, A.; Furnari, B.; Mondesert, O.; Russell, P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 1999, 397, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, H.; Liu, Y.-C.; Jelinek, T.; Zhang, L.; Ruoslahti, E.; Fu, H. Isolation of High-Affinity Peptide Antagonists of 14-3-3 Proteins by Phage Display. Biochemistry 1999, 38, 12499–12504. [Google Scholar] [CrossRef] [PubMed]
- Petosa, C.; Masters, S.C.; Bankston, L.A.; Pohl, J.; Wang, B.; Fu, H.; Liddington, R.C. 14-3-3zeta binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem. 1998, 273, 16305–16310. [Google Scholar] [CrossRef] [PubMed]
- Glas, A.; Bier, D.; Hahne, G.; Rademacher, C.; Ottmann, C.; Grossmann, T.N. Constrained peptides with target-adapted cross-links as inhibitors of a pathogenic protein-protein interaction. Angew. Chem. Int. Ed. Engl. 2014, 53, 2489–2493. [Google Scholar] [CrossRef] [PubMed]
- Cromm, P.M.; Wallraven, K.; Glas, A.; Bier, D.; Fürstner, A.; Ottmann, C.; Grossmann, T.N. Constraining an Irregular Peptide Secondary Structure through Ring-Closing Alkyne Metathesis. Chembiochem 2016, 17, 1915–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milroy, L.-G.; Bartel, M.; Henen, M.A.; Leysen, S.; Adriaans, J.M.C.; Brunsveld, L.; Landrieu, I.; Ottmann, C. Stabilizer-Guided Inhibition of Protein-Protein Interactions. Angew. Chem. Int. Ed. Engl. 2015, 54, 15720–15724. [Google Scholar] [CrossRef] [PubMed]
- Corradi, V.; Mancini, M.; Santucci, M.A.; Carlomagno, T.; Sanfelice, D.; Mori, M.; Vignaroli, G.; Falchi, F.; Manetti, F.; Radi, M.; et al. Computational techniques are valuable tools for the discovery of protein-protein interaction inhibitors: The 14-3-3σ case. Bioorg. Med. Chem. Lett. 2011, 21, 6867–6871. [Google Scholar] [CrossRef] [PubMed]
- Thiel, P.; Röglin, L.; Meissner, N.; Hennig, S.; Kohlbacher, O.; Ottmann, C. Virtual screening and experimental validation reveal novel small-molecule inhibitors of 14-3-3 protein-protein interactions. Chem. Commun. Camb. Engl. 2013, 49, 8468–8470. [Google Scholar] [CrossRef] [PubMed]
- Sijbesma, E.; Skora, L.; Leysen, S.; Brunsveld, L.; Koch, U.; Nussbaumer, P.; Jahnke, W.; Ottmann, C. Identification of Two Secondary Ligand Binding Sites in 14-3-3 Proteins Using Fragment Screening. Biochemistry 2017, 56, 3972–3982. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples not available. |





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ballone, A.; Centorrino, F.; Ottmann, C. 14-3-3: A Case Study in PPI Modulation. Molecules 2018, 23, 1386. https://doi.org/10.3390/molecules23061386
Ballone A, Centorrino F, Ottmann C. 14-3-3: A Case Study in PPI Modulation. Molecules. 2018; 23(6):1386. https://doi.org/10.3390/molecules23061386
Chicago/Turabian StyleBallone, Alice, Federica Centorrino, and Christian Ottmann. 2018. "14-3-3: A Case Study in PPI Modulation" Molecules 23, no. 6: 1386. https://doi.org/10.3390/molecules23061386




