Microwave Assisted Sol-Gel Synthesis of Silica-Spider Silk Composites
Abstract
:1. Introduction
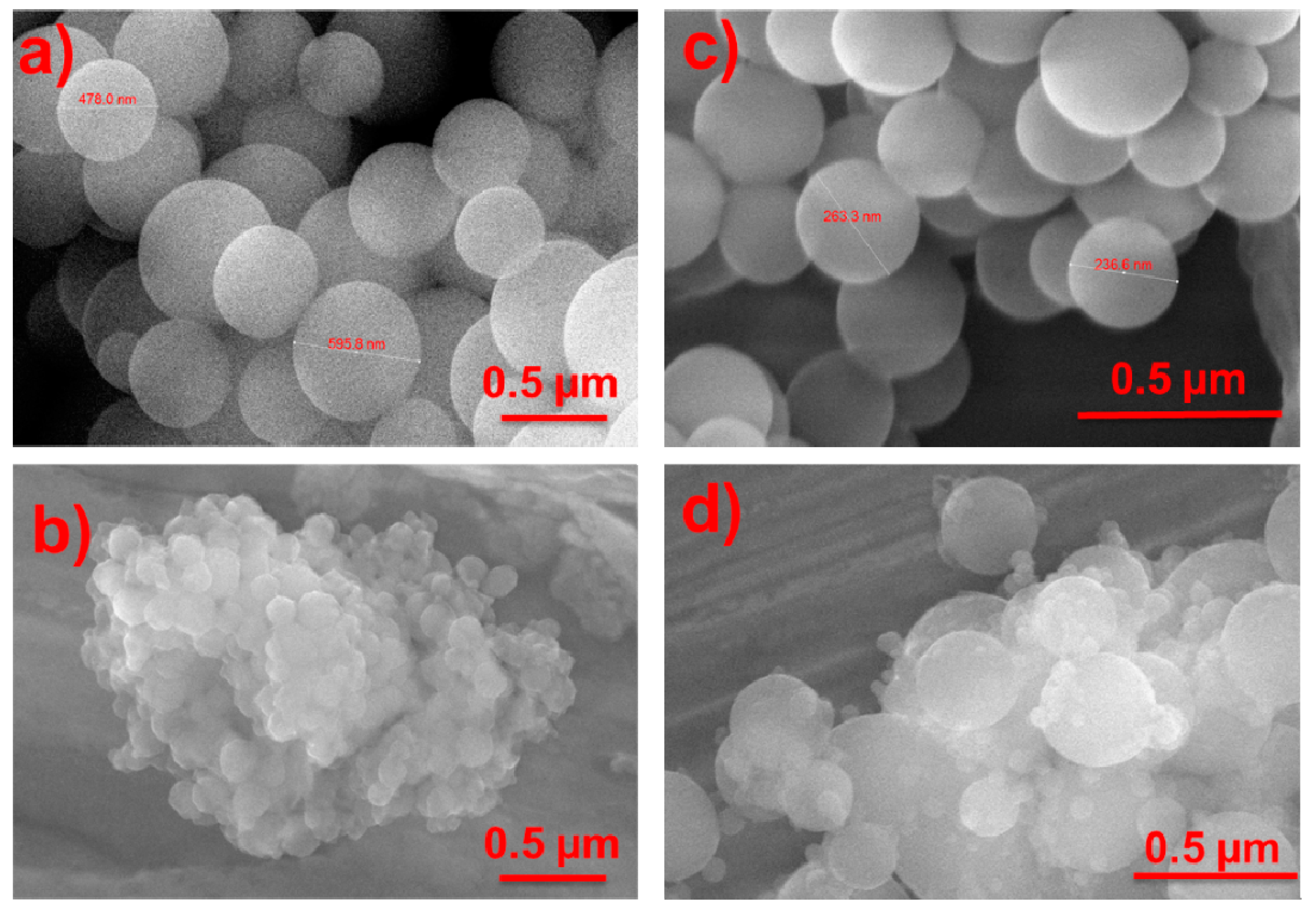
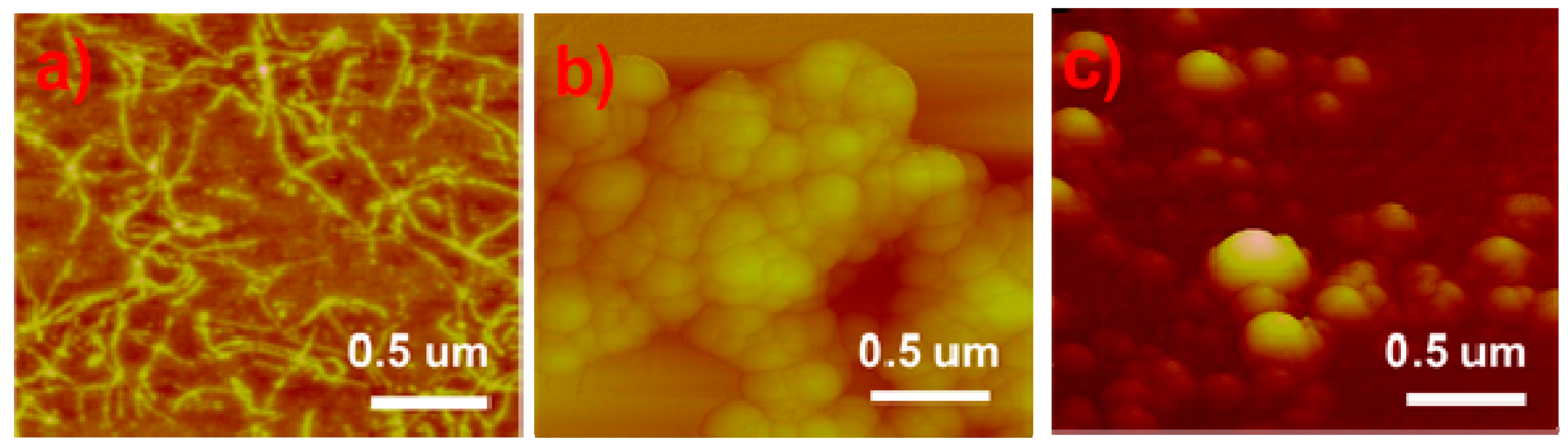
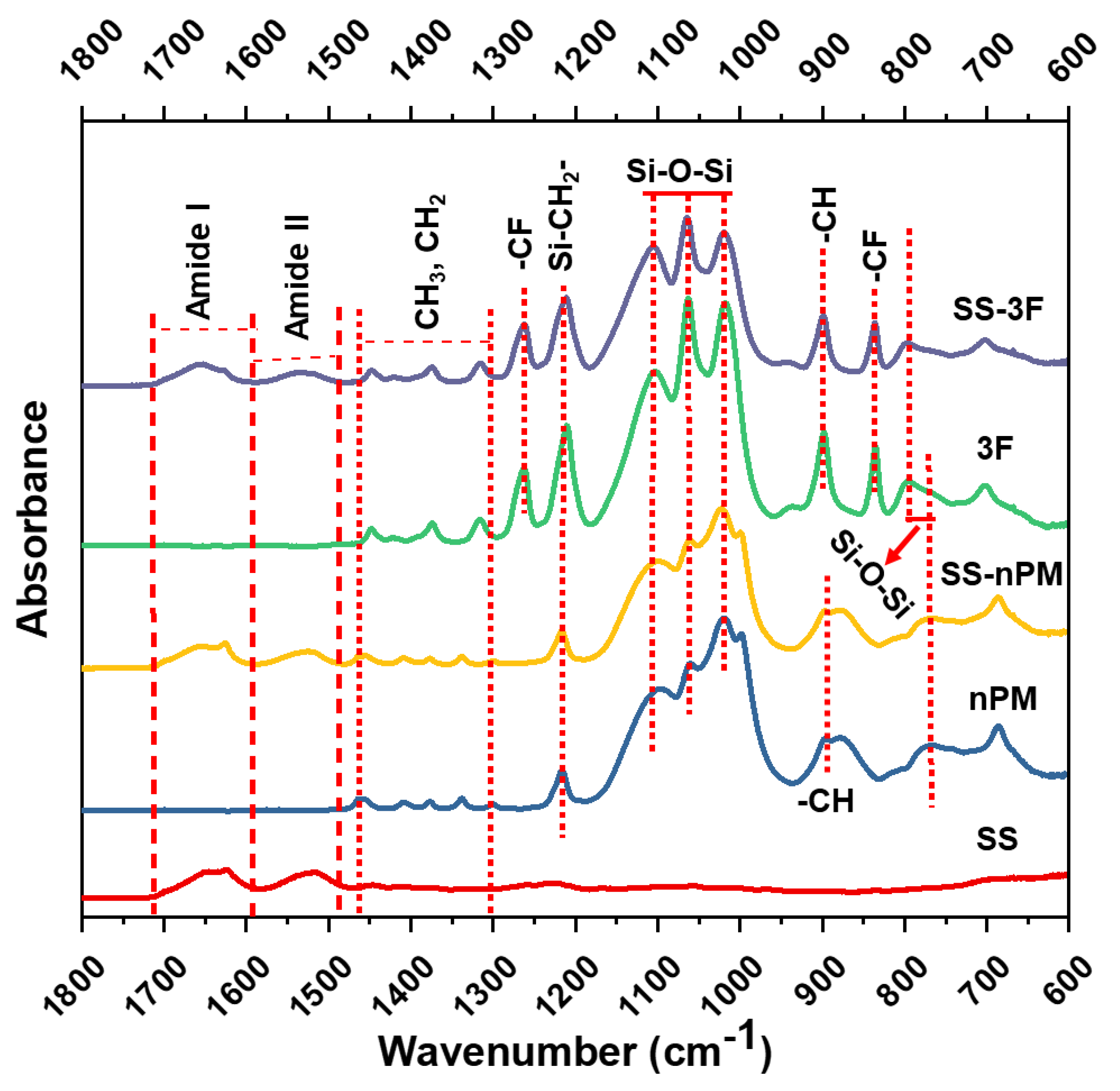
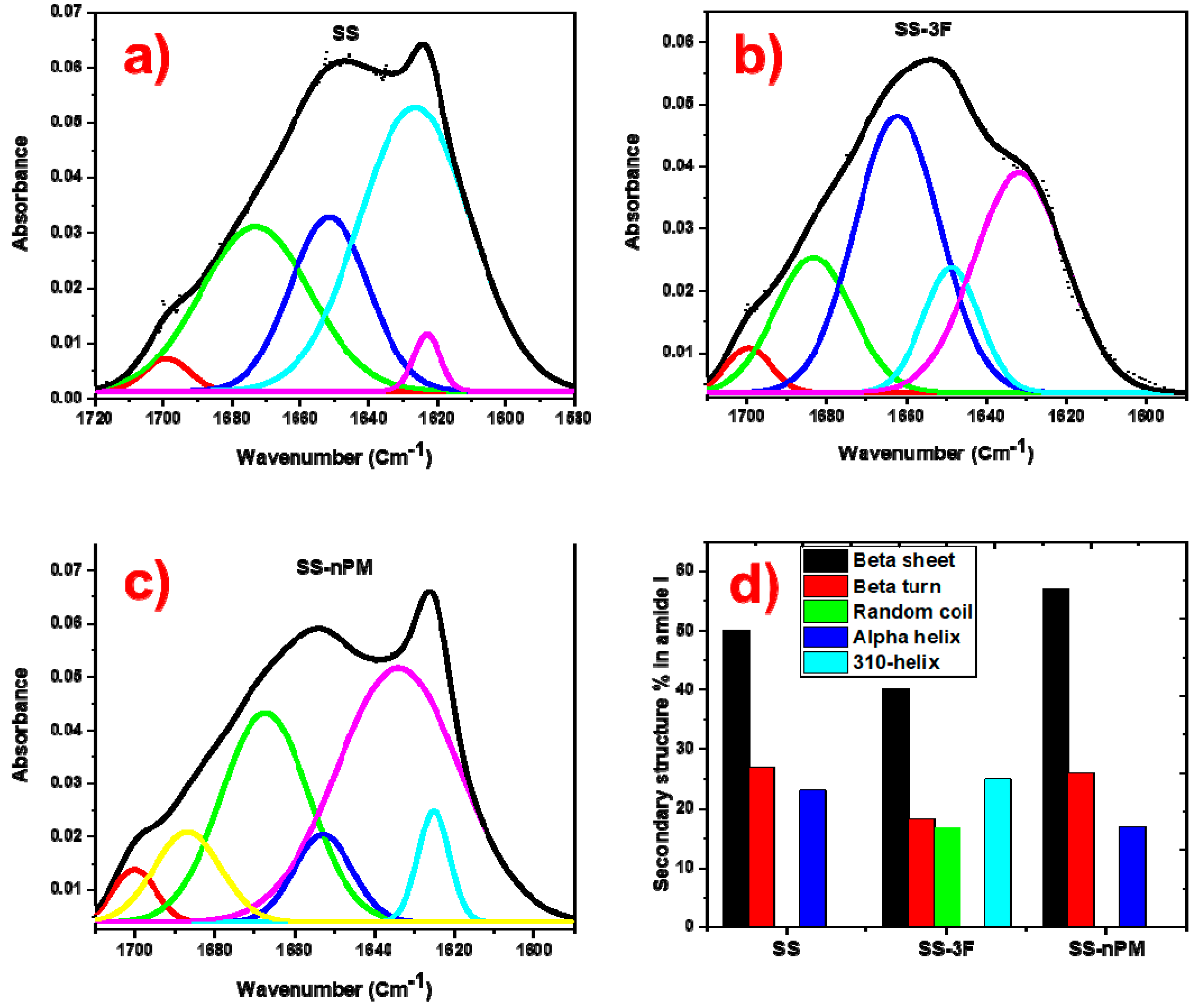
2. Results and Discussion
3. Materials and Methods
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tan, W.; Wang, K.; He, X.; Zhao, X.J.; Drake, T.; Wang, L.; Bagwe, R.P. Bionanotechnology based on silica nanoparticles. Med. Res. Rev. 2004, 24, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Coradin, T.; Livage, J. Aqueous Silicates in Biological Sol–Gel Applications: New Perspectives for Old Precursors. Acc. Chem. Res. 2007, 40, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I.A.; Padavettan, V. Synthesis of Silica nanoparticles by Sol-Gel: Size-dependent properties, surface modification, and applications in silica-polymer nanocompositesa review. J. Nanomater. 2012, 8, 1–15. [Google Scholar] [CrossRef]
- Tripathi, V.S.; Kandimalla, V.B.; Ju, H. Preparation of ormosil and its applications in the immobilizing biomolecules. Sens. Actuators B Chem. 2006, 114, 1071–1082. [Google Scholar] [CrossRef]
- Ronda, L.; Bettati, S.; Bruno, S. Immobilization of Proteins in Ormosil Gels: Functional Properties and Applications. Curr. Org. Chem. 2015, 19, 1677–1683. [Google Scholar] [CrossRef]
- Bottini, M.; Di Venere, A.; Lugli, P.; Rosato, N. Conformation and stability of myoglobin in dilute and crowded organically modified media. J. Non. Cryst. Solids 2004, 343, 101–108. [Google Scholar] [CrossRef]
- Albarran, B.; To, R.; Stayton, P.S. A TAT–streptavidin fusion protein directs uptake of biotinylated cargo into mammalian cells. Protein Eng. Des. Sel. 2005, 18, 147–152. [Google Scholar] [CrossRef]
- Asai, T.; Trinh, R.; Ng, P.P.; Penichet, M.L.; Wims, L.A.; Morrison, S.L. A human biotin acceptor domain allows site-specific conjugation of an enzyme to an antibody-avidin fusion protein for targeted drug delivery. Biomol. Eng. 2005, 21, 145–155. [Google Scholar] [CrossRef] [Green Version]
- Icke, C. Fusion Proteins with Anticoagulant and Fibrinolytic Properties: Functional Studies and Structural Considerations. Mol. Pharm. 2002, 62, 203–209. [Google Scholar] [CrossRef] [Green Version]
- Park, E.; Starzyk, R.M.; McGrath, J.P.; Lee, T.; George, J.; Schutz, A.J.; Lynch, P.; Putney, S.D. Production and Characterization of Fusion Proteins Containing Transferrin and Nerve Growth Factor. J. Drug Target. 1998, 6, 53–64. [Google Scholar] [CrossRef]
- Dubrovsky, T.; Tronin, A.; Dubrovskaya, S.; Guryev, O.; Nicolini, C. Langmuir films of Fc binding receptors engineered from protein A and protein G as a sublayer for immunoglobulin orientation. Thin Solid Films 1996, 284–285, 698–702. [Google Scholar] [CrossRef]
- Frey, W.; Meyer, D.E.; Chilkoti, A. Dynamic Addressing of a Surface Pattern by a Stimuli-Responsive Fusion Protein. Adv. Mater. 2003, 15, 248–251. [Google Scholar] [CrossRef]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435. [Google Scholar] [CrossRef] [PubMed]
- Slocik, J.M.; Naik, R.R.; Stone, M.O.; Wright, D.W. Viral templates for gold nanoparticle synthesis. J. Mater. Chem. 2005, 15, 749–753. [Google Scholar] [CrossRef]
- Padilla, J.E.; Colovos, C.; Yeates, T.O. Nanohedra: Using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl. Acad. Sci. USA 2001, 98, 2217–2221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paternolli, C.; Ghisellini, P.; Nicolini, C. Pollutant sensing layer based on cytochrome P450. Mater. Sci. Eng. C 2002, 22, 155–159. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Schuster, B.; Pum, D. Nanotechnology and biomimetics with 2-D protein crystals. IEEE Eng. Med. Biol. Mag. 2003, 22, 140–150. [Google Scholar] [CrossRef]
- Mieszawska, A.J.; Fourligas, N.; Georgakoudi, I.; Ouhib, N.M.; Belton, D.J.; Perry, C.C.; Kaplan, D.L. Osteoinductive silk-silica composite biomaterials for bone regeneration. Biomaterials 2010, 31, 8902–8910. [Google Scholar] [CrossRef]
- Teulé, F.; Addison, B.; Cooper, A.R.; Ayon, J.; Henning, R.W.; Benmore, C.J.; Holland, G.P.; Yarger, J.L.; Lewis, R.V. Combining flagelliform and dragline spider silk motifs to produce tunable synthetic biopolymer fibers. Biopolymers 2012, 97, 418–431. [Google Scholar] [CrossRef]
- An, B.; Jenkins, J.E.; Sampath, S.; Holland, G.P.; Hinman, M.; Yarger, J.L.; Lewis, R. Reproducing natural spider silks’ copolymer behavior in synthetic silk mimics. Biomacromolecules 2012, 13, 3938–3948. [Google Scholar] [CrossRef]
- Xia, X.X.; Qian, Z.G.; Ki, C.S.; Park, Y.H.; Kaplan, D.L.; Lee, S.Y. Native-sized recombinant spider silk protein produced in metabolically engineered Escherichia coli results in a strong fiber. Proc. Natl. Acad. Sci. USA 2010, 107, 14059–14063. [Google Scholar] [CrossRef] [PubMed]
- Minoura, N.; Aiba, S.I.; Gotoh, Y.; Tsukada, M.; Imai, Y. Attachment and growth of cultured fibroblast cells on silk protein matrices. J. Biomed. Mater. Res. 1995, 29, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.W.; Roskov, K.E.; Lin-Gibson, S.; Kaplan, D.L.; Becker, M.L.; Simon, C.G. Characterization and optimization of RGD-containing silk blends to support osteoblastic differentiation. Biomaterials 2008, 29, 2556–2563. [Google Scholar] [CrossRef] [PubMed]
- Sofia, S.; McCarthy, M.B.; Gronowicz, G.; Kaplan, D.L. Functionalized silk-based biomaterials for bone formation. J. Biomed. Mater. Res. 2001, 54, 139–148. [Google Scholar] [CrossRef]
- Baoyong, L.; Jian, Z.; Denglong, C.; Min, L. Evaluation of a new type of wound dressing made from recombinant spider silk protein using rat models. Burns 2010, 36, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Hijirida, D.H.; Do, K.G.; Michal, C.; Wong, S.; Zax, D.; Jelinski, L.W. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 1996, 71, 3442–3447. [Google Scholar] [CrossRef] [Green Version]
- Gomes, S.C.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. Antimicrobial functionalized genetically engineered spider silk. Biomaterials 2011, 32, 4255–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teulé, F.; Cooper, A.R.; Furin, W.A.; Bittencourt, D.; Rech, E.L.; Brooks, A.; Lewis, R.V. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat. Protoc. 2009, 4, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Albertson, A.E.; Teulé, F.; Weber, W.; Yarger, J.L.; Lewis, R.V. Effects of different post-spin stretching conditions on the mechanical properties of synthetic spider silk fibers. J. Mech. Behav. Biomed. Mater. 2014, 29, 225–234. [Google Scholar] [CrossRef]
- Gomes, S.; Gallego-Llamas, J.; Leonor, I.B.; Mano, J.F.; Reis, R.L.; Kaplan, D.L. In Vivo Biological Responses to Silk Proteins Functionalized with Bone Sialoprotein. Macromol. Biosci. 2013, 13, 444–454. [Google Scholar] [CrossRef]
- Slotta, U.; Tammer, M.; Kremer, F.; Koelsch, P.; Scheibel, T. Structural analysis of spider silk films. Supramol. Chem. 2006, 18, 465–471. [Google Scholar] [CrossRef]
- Huemmerich, D.; Slotta, U.; Scheibel, T. Processing and modification of films made from recombinant spider silk proteins. Appl. Phys. A Mater. Sci. Process. 2006, 82, 219–222. [Google Scholar] [CrossRef]
- Krishnaji, S.T.; Huang, W.; Rabotyagova, O.; Kharlampieva, E.; Choi, I.; Tsukruk, V.V.; Naik, R.; Cebe, P.; Kaplan, D.L. Thin Film Assembly of Spider Silk-like Block Copolymers. Langmuir 2011, 27, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Leal-Egaña, A.; Scheibel, T.R. Engineered spider silk protein-based composites for drug delivery. Macromol. Biosci. 2013, 13, 1431–1437. [Google Scholar] [CrossRef] [PubMed]
- Tucker, C.L.; Jones, J.A.; Bringhurst, H.N.; Copeland, C.G.; Addison, J.B.; Weber, W.S.; Mou, Q.; Yarger, J.L.; Lewis, R.V. Mechanical and physical properties of recombinant spider silk films using organic and aqueous solvents. Biomacromolecules 2014, 15, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, C.; Belleville, P.; Popall, M.; Nicole, L. Applications of advanced hybrid organic-inorganic nanomaterials: From laboratory to market. Chem. Soc. Rev. 2011, 40, 696–753. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Turner, N.W.; Britt, D.W. Trifluorosilane induced structural transitions in beta-lactoglobulin in sol and gel. Colloids Surf. B Biointerfaces 2014, 119, 6–13. [Google Scholar] [CrossRef]
- Pereira, C.; Alves, C.; Monteiro, A.; Magén, C.; Pereira, A.M.; Ibarra, A.; Ibarra, M.R.; Tavares, P.B.; Araújo, J.P.; Blanco, G.; et al. Designing novel hybrid materials by one-pot co-condensation: From hydrophobic mesoporous silica nanoparticles to superamphiphobic cotton textiles. ACS Appl. Mater. Interfaces 2011, 3, 2289–2299. [Google Scholar] [CrossRef]
- Suegama, P.H.; Aoki, I.V. Electrochemical behavior of carbon steel pre-treated with an organo functional bis-silane filled with copper phthalocyanine. J. Braz. Chem. Soc. 2008, 19, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Lenza, R.F.S.; Vasconcelos, W.L. Structural evolution of silica sols modified with formamide. Mater. Res. 2001, 4, 175–179. [Google Scholar] [CrossRef]
- Kim, M.T. Deposition behavior of hexamethydisiloxane films based on the FTIR analysis of Si–O–Si and Si–CH3 bonds. Thin Solid Films 1997, 311, 157–163. [Google Scholar] [CrossRef]
- Harris, T.I.; Gaztambide, D.A.; Day, B.A.; Brock, C.L.; Ruben, A.L.; Jones, J.A.; Lewis, R.V. Sticky Situation: An Investigation of Robust Aqueous-Based Recombinant Spider Silk Protein Coatings and Adhesives. Biomacromolecules 2016, 17, 3761–3772. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the SS, SS-nPM, and SS-3F are available from the authors. |

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giasuddin, A.B.M.; Britt, D.W. Microwave Assisted Sol-Gel Synthesis of Silica-Spider Silk Composites. Molecules 2019, 24, 2521. https://doi.org/10.3390/molecules24142521
Giasuddin ABM, Britt DW. Microwave Assisted Sol-Gel Synthesis of Silica-Spider Silk Composites. Molecules. 2019; 24(14):2521. https://doi.org/10.3390/molecules24142521
Chicago/Turabian StyleGiasuddin, Abul Bashar Mohammad, and David W. Britt. 2019. "Microwave Assisted Sol-Gel Synthesis of Silica-Spider Silk Composites" Molecules 24, no. 14: 2521. https://doi.org/10.3390/molecules24142521






