A Sample Preparation Technique Using Biocompatible Composites for Biomedical Applications
Abstract
:1. Introduction
2. Results and Discussion
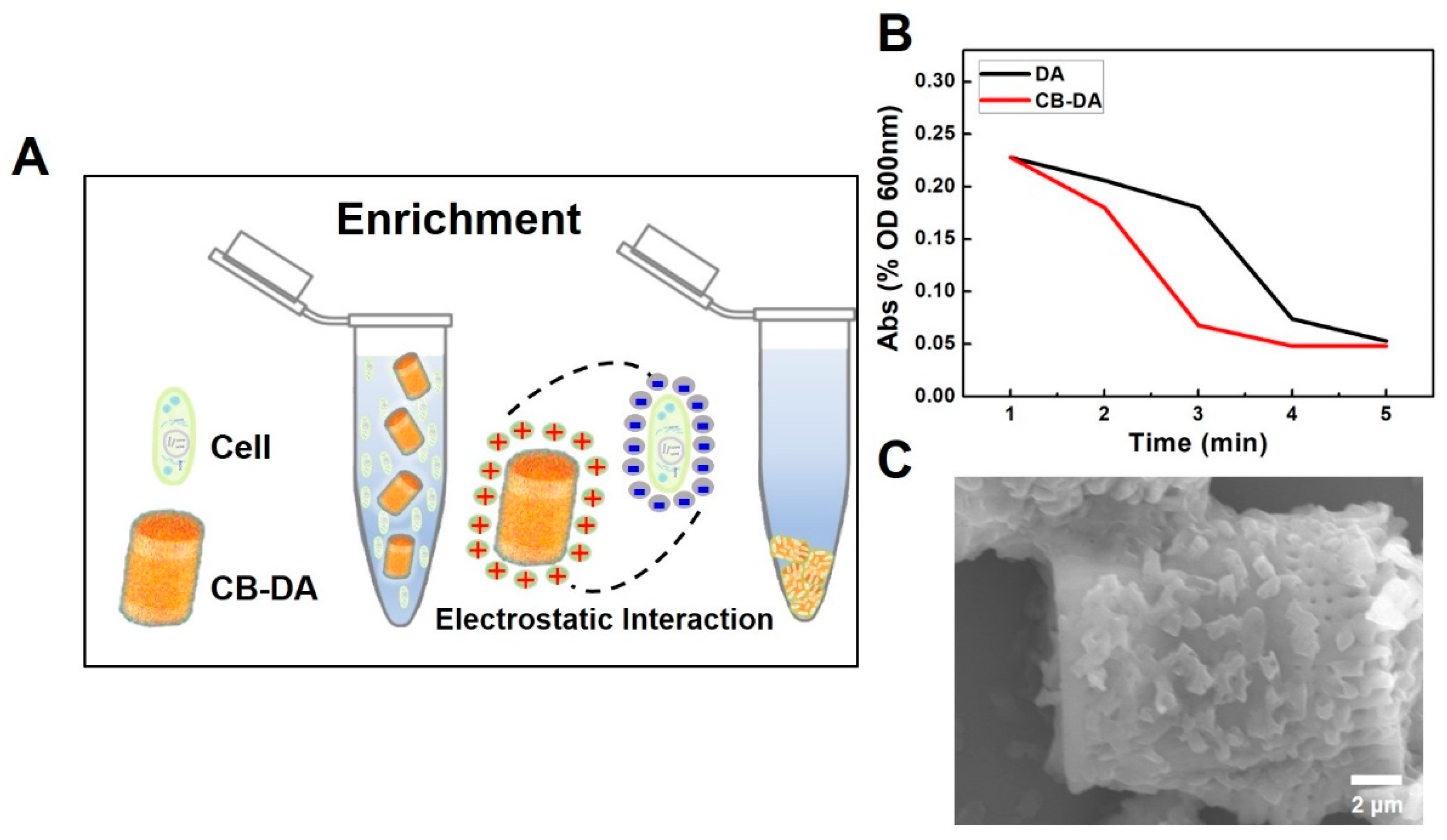
2.1. Design and Principle of the CB–DA Biocompatible Composite
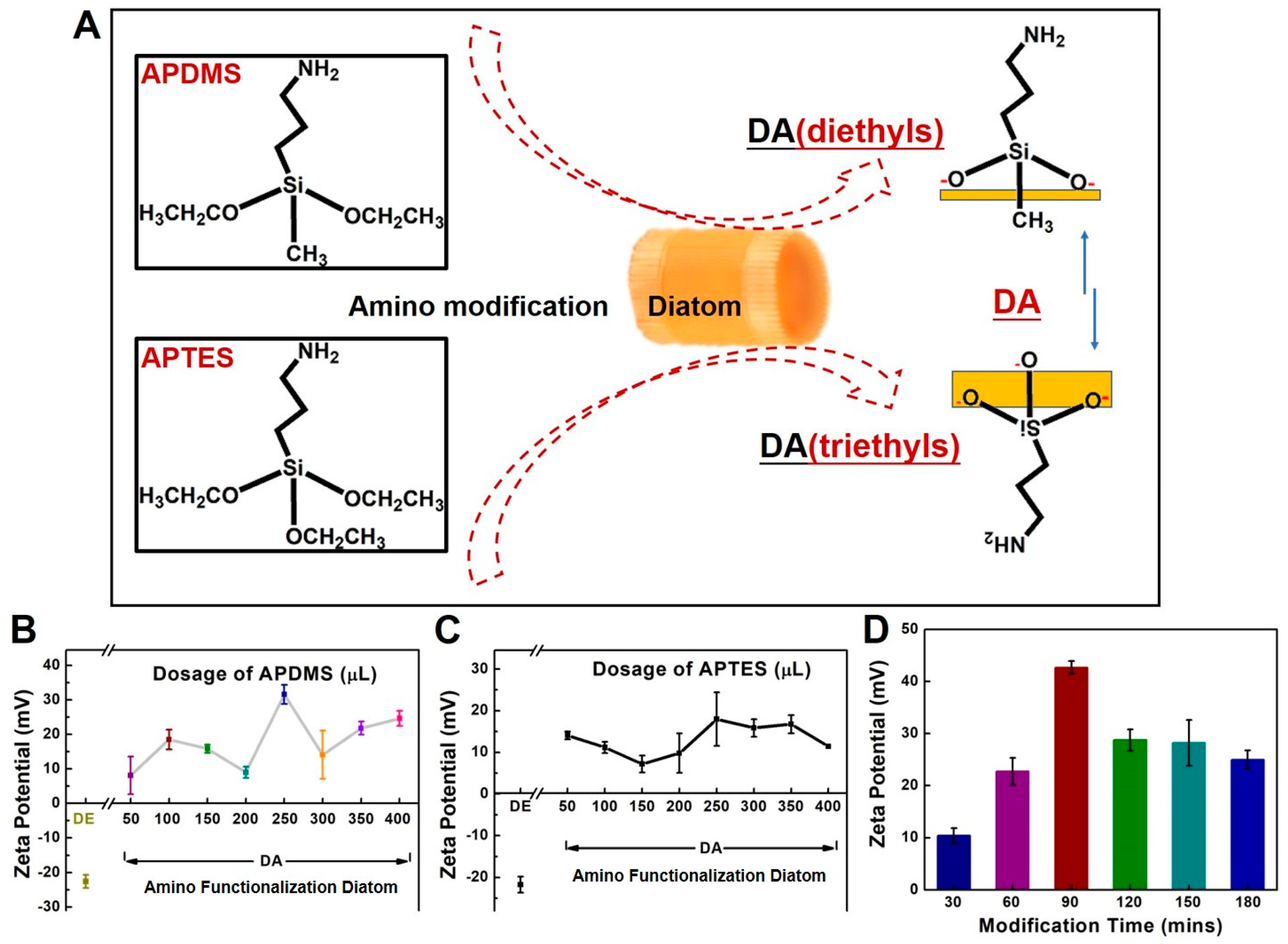
2.2. Preparation and Characterization of DA
2.3. Preparation and Characterization of CB–DA Biocompatible Composites
2.4. Cell and Pathogen Enrichment Using the Biocompatible Composite
2.5. Nucleic Acid Isolation Using the Biocompatible Composite
3. Experimental Section
3.1. Chemicals and Reagents
3.2. Biological Samples
3.3. Preparation of Biocompatible Composites
3.4. Characterization of the Biocompatible Composites
3.5. Cell and Pathogen Enrichment and Nucleic Acid Capturing
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Koo, B.; Hong, K.H.; Jin, C.E.; Kim, J.Y.; Kim, S.-H.; Shin, Y. Arch-shaped multiple-target sensing for rapid diagnosis and identification of emerging infectious pathogens. Biosens. Bioelectron. 2018, 119, 79–85. [Google Scholar] [CrossRef]
- Hahm, B.-K.; Kim, H.; Singh, A.K.; Bhunia, A.K. Pathogen enrichment device (PED) enables one-step growth, enrichment and separation of pathogen from food matrices for detection using bioanalytical platforms. J. Microbiol. Methods 2015, 117, 64–73. [Google Scholar] [CrossRef]
- Zhao, F.; Koo, B.; Liu, H.; Jin, C.E.; Shin, Y. A single-tube approach for in vitro diagnostics using diatomaceous earth and optical sensor. Biosens. Bioelectron. 2018, 99, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.E.; Koo, B.; Lee, T.Y.; Han, K.; Lim, S.B.; Park, I.J.; Shin, Y. Simple and low-cost sampling of cell-free nucleic acids from blood plasma for rapid and sensitive detection of circulating tumor DNA. Adv. Sci. 2018, 5, 1800614. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, S.; Koh, Y.-S.; Kim, Y.; Li, W. Isolation and Identification of Benzochroman and Acylglycerols from Massa Medicata Fermentata and Their Inhibitory Effects on LPS-Stimulated Cytokine Production in Bone Marrow-Derived Dendritic Cells. Molecules 2018, 23, 2400. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, F.; Jin, C.E.; Koo, B.; Lee, E.Y.; Zhong, L.; Yun, K.; Shin, Y. Large Instrument-and Detergent-Free Assay for Ultrasensitive Nucleic Acids Isolation via Binary Nanomaterial. Anal. Chem. 2018, 90, 5108–5115. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shen, C.; Huang, Y.-X.; Li, Y.-N.; Liu, X.-F.; Liu, X.-M.; Liu, J.-H. A New Strategy for Rapidly Screening Natural Inhibitors Targeting the PCSK9/LDLR Interaction In Vitro. Molecules 2018, 23, 2397. [Google Scholar] [CrossRef]
- Jin, C.E.; Koo, B.; Lee, E.Y.; Kim, J.E.; Kim, S.H.; Shin, Y. Simple and label-free pathogen enrichment via homobifunctional imidoesters using a microfluidic (SLIM) system for ultrasensitive pathogen detection in various clinical specimens. Biosens. Bioelectron. 2018, 111, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Sim, J.; Oh, K.; Sung, G.; Lee, A.; Shrinidhi, A.; Thirunarayanan, A.; Shetty, D.; Kim, K. Enrichment of specifically labeled proteins by an immobilized host molecule. Angew. Chem. Int. Ed. 2017, 56, 2395–2398. [Google Scholar] [CrossRef] [PubMed]
- Barooah, N.; Kunwar, A.; Khurana, R.; Bhasikuttan, A.C.; Mohanty, J. Stimuli-Responsive Cucurbit [7] uril-Mediated BSA Nanoassembly for Uptake and Release of Doxorubicin. Chem. Asian J. 2017, 12, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luan, Y.; Koo, B.; Lee, E.Y.; Joo, J.; Dao, T.N.T.; Zhao, F.; Zhong, L.; Yun, K.; Shin, Y. Cucurbituril-based Reusable Nanocomposites for Efficient Molecular Encapsulation. ACS Sustain. Chem. Eng. 2019, 7, 5440–5448. [Google Scholar] [CrossRef]
- Lu, H.-T. Synthesis and characterization of amino-functionalized silica nanoparticles. Colloid J. 2013, 75, 311–318. [Google Scholar] [CrossRef]
- Kim, K.; Selvapalam, N.; Ko, Y.H.; Park, K.M.; Kim, D.; Kim, J. Functionalized cucurbiturils and their applications. Chem. Soci. Rev. 2007, 36, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, J.; Xue, S.-F.; Tao, Z.; Zhu, Q.-J.; Tang, Q. Determination of thiabendazole in aqueous solutions using a cucurbituril-enhanced fluorescence method. J. Incl. Phenom. Macrocyclic Chem. 2012, 72, 397–404. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, H.; Bai, H.; Bai, Y.; Wang, S.; Zhang, X. Polypseudorotaxane constructed from cationic polymer with cucurbit [7] uril for controlled antibacterial activity. ACS Macro Lett. 2016, 5, 1109–1113. [Google Scholar] [CrossRef]
- Chao, C.-L.; Huang, H.-W.; Huang, H.-C.; Chao, H.-F.; Yu, S.-W.; Su, M.-H.; Wang, C.-J.; Lin, H.-C. Inhibition of Amyloid Beta Aggregation and Deposition of Cistanche tubulosa Aqueous Extract. Molecules 2019, 24, 687. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Liu, F.; Wang, Z.; Li, G.; Karamanos, Y. Detection of tumor invasive biomarker using a peptamer of signal conversion and signal amplification. Anal. Chem. 2016, 88, 3662–3668. [Google Scholar] [CrossRef]
- Lee, J.W.; Samal, S.; Selvapalam, N.; Kim, H.-J.; Kim, K. Cucurbituril homologues and derivatives: New opportunities in supramolecular chemistry. Acc. Chem. Res. 2003, 36, 621–630. [Google Scholar] [CrossRef]
- Xiao, X.; Li, W.; Jiang, J. Porphyrin-cucurbituril organic molecular porous material: Structure and iodine adsorption properties. Inorg. Chem. Commun. 2013, 35, 156–159. [Google Scholar] [CrossRef]
- Pennakalathil, J.; Jahja, E.; Özdemir, E.S.; Konu, O.; Tuncel, D. Red emitting, cucurbituril-capped, pH-responsive conjugated oligomer-based nanoparticles for drug delivery and cellular imaging. Biomacromolecules 2014, 15, 3366–3374. [Google Scholar] [CrossRef]
- Zhou, X.; Su, X.; Pathak, P.; Vik, R.; Vinciguerra, B.; Isaacs, L.; Jayawickramarajah, J. Host–Guest Tethered DNA Transducer: ATP Fueled Release of a Protein Inhibitor from Cucurbit [7] uril. J. Am. Chem. Soc. 2017, 139, 13916–13921. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Lee, E.Y.; Shin, Y. Improved Reversible Cross-Linking-Based Solid-Phase RNA Extraction for Pathogen Diagnostics. Anal. Chem. 2018, 90, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- El-Sheshtawy, H.S.; Chatterjee, S.; Assaf, K.I.; Shinde, M.N.; Nau, W.M.; Mohanty, J. A Supramolecular Approach for Enhanced Antibacterial Activity and Extended Shelf-life of Fluoroquinolone Drugs with Cucurbit [7] uril. Sci. Rep. 2018, 8, 13925. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.; Zeng, X.; Gao, Y.; Qiao, S.; Zhang, X.; Xu, J.; Liu, J. Cucurbituril as a Versatile Tool to Tune the Functions of Proteins. Isr. J. Chem. 2018, 58, 286–295. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |



| Primer | Sequences (5′→3′) | Annealing Temp. (°C) | |
|---|---|---|---|
| E coli | Ecoli-rodA-195F | GCA AAC CAC CTT TGG TCG | 58 |
| Ecoli-rodA-195R | CTG TGG GTG TGG ATT GAC AT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Noh, G.S.; Luan, Y.; Qiao, Z.; Koo, B.; Jang, Y.O.; Shin, Y. A Sample Preparation Technique Using Biocompatible Composites for Biomedical Applications. Molecules 2019, 24, 1321. https://doi.org/10.3390/molecules24071321
Liu H, Noh GS, Luan Y, Qiao Z, Koo B, Jang YO, Shin Y. A Sample Preparation Technique Using Biocompatible Composites for Biomedical Applications. Molecules. 2019; 24(7):1321. https://doi.org/10.3390/molecules24071321
Chicago/Turabian StyleLiu, Huifang, Geun Su Noh, Yange Luan, Zhen Qiao, Bonhan Koo, Yoon Ok Jang, and Yong Shin. 2019. "A Sample Preparation Technique Using Biocompatible Composites for Biomedical Applications" Molecules 24, no. 7: 1321. https://doi.org/10.3390/molecules24071321





