Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism
Abstract
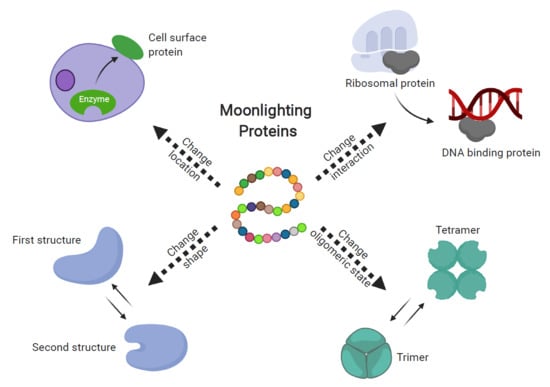
:1. Introduction
2. Examples of Moonlighting Proteins and Factors that Affect Function
2.1. Cellular Localization
2.2. Interactions with Other Proteins and Molecules
2.3. Environmental Stress
2.4. Changes in Protein Structure
2.4.1. Intrinsically Disordered Proteins
2.4.2. Metamorphic Proteins
2.4.3. Morpheein Proteins
3. Moonlighting Proteins in Cellular Complexity
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jeffery, C.J. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Jeffery, C.J. Protein moonlighting: What is it, and why is it important? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.; Chen, C.; Amblee, V.; Liu, H.; Mathur, T.; Zwicke, G.; Zabad, S.; Patel, B.; Thakkar, J.; Jeffery, C.J. MoonProt: A database for proteins that are known to moonlight. Nucleic Acids Res. 2015, 43, D277–D282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zabad, S.; Liu, H.; Wang, W.; Jeffery, C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 2018, 46, D640–D644. [Google Scholar] [CrossRef] [Green Version]
- Gentili, P.L. The Fuzziness of the Molecular World and Its Perspectives. Molecules 2018, 23, 2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuxreiter, M. Towards a Stochastic Paradigm: From Fuzzy Ensembles to Cellular Functions. Molecules 2018, 23, 3008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, B.; Martin, A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011, 79, 3476–3491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffery, C. Intracellular proteins moonlighting as bacterial adhesion factors. Aims Microbiol. 2018, 4, 362–376. [Google Scholar] [CrossRef]
- Wang, W.; Jeffery, C.J. An analysis of surface proteomics results reveals novel candidates for intracellular/surface moonlighting proteins in bacteria. Mol. Biosyst. 2016, 12, 1420–1431. [Google Scholar] [CrossRef]
- Kainulainen, V.; Korhonen, T.K. Dancing to another tune-adhesive moonlighting proteins in bacteria. Biology 2014, 3, 178–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sha, J.; Erova, T.E.; Alyea, R.A.; Wang, S.; Olano, J.P.; Pancholi, V.; Chopra, A.K. Surface-expressed enolase contributes to the pathogenesis of clinical isolate SSU of Aeromonas hydrophila. J. Bacteriol. 2009, 191, 3095–3107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, S.; Kulshreshtha, P.; Bambah Mukku, D.; Bhatnagar, R. α-Enolase binds to human plasminogen on the surface of Bacillus anthracis. Biochim. Biophys. Acta 2008, 1784, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Knaust, A.; Weber, M.V.; Hammerschmidt, S.; Bergmann, S.; Frosch, M.; Kurzai, O. Cytosolic proteins contribute to surface plasminogen recruitment of Neisseria meningitidis. J. Bacteriol. 2007, 189, 3246–3255. [Google Scholar] [CrossRef] [Green Version]
- Kolberg, J.; Aase, A.; Bergmann, S.; Herstad, T.K.; Rodal, G.; Frank, R.; Rohde, M.; Hammerschmidt, S. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 2006, 152, 1307–1317. [Google Scholar] [CrossRef] [Green Version]
- Mundodi, V.; Kucknoor, A.S.; Alderete, J.F. Immunogenic and plasminogen-binding surface-associated alpha-enolase of Trichomonas vaginalis. Infect. Immun. 2008, 76, 523–531. [Google Scholar] [CrossRef] [Green Version]
- Antikainen, J.; Kuparinen, V.; Lahteenmaki, K.; Korhonen, T.K. Enolases from Gram-positive bacterial pathogens and commensal lactobacilli share functional similarity in virulence-associated traits. Fems Immunol. Med. Microbiol. 2007, 51, 526–534. [Google Scholar] [CrossRef] [Green Version]
- Collen, D.; Verstraete, M. Molecular biology of human plasminogen. II. Metabolism in physiological and some pathological conditions in man. Thromb. Diath. Haemorrh. 1975, 34, 403–408. [Google Scholar]
- Dano, K.; Andreasen, P.A.; Grondahl-Hansen, J.; Kristensen, P.; Nielsen, L.S.; Skriver, L. Plasminogen activators, tissue degradation, and cancer. Adv. Cancer Res. 1985, 44, 139–266. [Google Scholar] [CrossRef]
- Esgleas, M.; Li, Y.; Hancock, M.A.; Harel, J.; Dubreuil, J.D.; Gottschalk, M. Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 2008, 154, 2668–2679. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, C.R.; Postol, E.; Nomizo, R.; Reis, L.F.; Brentani, R.R. Identification of enolase as a laminin-binding protein on the surface of Staphylococcus aureus. Microbes Infect. 2004, 6, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, H.; Uchida, H.; Kawai, Y.; Kawasaki, T.; Wakahara, N.; Matsuo, H.; Watanabe, M.; Kitazawa, H.; Ohnuma, S.; Miura, K.; et al. Cell surface Lactobacillus plantarum LA 318 glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin. J. Appl. Microbiol. 2008, 104, 1667–1674. [Google Scholar] [CrossRef]
- Patel, D.K.; Shah, K.R.; Pappachan, A.; Gupta, S.; Singh, D.D. Cloning, expression and characterization of a mucin-binding GAPDH from Lactobacillus acidophilus. Int. J. Biol. Macromol. 2016, 91, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Lottenberg, R.; Broder, C.C.; Boyle, M.D.; Kain, S.J.; Schroeder, B.L.; Curtiss, R., 3rd. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J. Bacteriol. 1992, 174, 5204–5210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pancholi, V.; Fischetti, V.A. A major surface protein on group a streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 1992, 176, 415–426. [Google Scholar] [CrossRef]
- Pancholi, V.; Fischetti, V.A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. USA 1993, 90, 8154–8158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terao, Y.; Yamaguchi, M.; Hamada, S.; Kawabata, S. Multifunctional glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pyogenes is essential for evasion from neutrophils. J. Biol. Chem. 2006, 281, 14215–14223. [Google Scholar] [CrossRef] [Green Version]
- Madureira, P.; Baptista, M.; Vieira, M.; Magalhaes, V.; Camelo, A.; Oliveira, L.; Ribeiro, A.; Tavares, D.; Trieu-Cuot, P.; Vilanova, M.; et al. Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 2007, 178, 1379–1387. [Google Scholar] [CrossRef] [Green Version]
- Xolalpa, W.; Vallecillo, A.J.; Lara, M.; Mendoza-Hernandez, G.; Comini, M.; Spallek, R.; Singh, M.; Espitia, C. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 2007, 7, 3332–3341. [Google Scholar] [CrossRef]
- Wang, Y.; Kelly, C.G.; Karttunen, J.T.; Whittall, T.; Lehner, P.J.; Duncan, L.; MacAry, P.; Younson, J.S.; Singh, M.; Oehlmann, W.; et al. CD40 is a cellular receptor mediating mycobacterial heat shock protein 70 stimulation of CC-chemokines. Immunity 2001, 15, 971–983. [Google Scholar] [CrossRef] [Green Version]
- Ehinger, S.; Schubert, W.D.; Bergmann, S.; Hammerschmidt, S.; Heinz, D.W. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: Crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 2004, 343, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Dupuy, J.; Volbeda, A.; Carpentier, P.; Darnault, C.; Moulis, J.M.; Fontecilla-Camps, J.C. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure 2006, 14, 129–139. [Google Scholar] [CrossRef]
- Walden, W.E.; Selezneva, A.I.; Dupuy, J.; Volbeda, A.; Fontecilla-Camps, J.C.; Theil, E.C.; Volz, K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science 2006, 314, 1903–1908. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Lee, W.; Kim, E.E. Crystal structures of human peroxiredoxin 6 in different oxidation states. Biochem. Biophys. Res. Commun. 2016, 477, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, F.; Tse, E.; Castro, H.; Makepeace, K.A.T.; Meinen, B.A.; Borchers, C.H.; Poole, L.B.; Bardwell, J.C.; Tomas, A.M.; Southworth, D.R.; et al. Chaperone activation and client binding of a 2-cysteine peroxiredoxin. Nat. Commun. 2019, 10, 659. [Google Scholar] [CrossRef] [PubMed]
- Sehnal, D.; Rose, A.S.; Kovca, J.; Burley, S.K.; Velankar, S. Mol*: Towards a common library and tools for web molecular graphics. In Workshop on Molecular Graphics and Visual Analysis of Molecular Data; Byška, J., Krone, M., Sommer, B., Eds.; The Eurographics Association: Aire-la-Ville, Switzerland, 2018; pp. 29–33. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Di Costanzo, L.; Christie, C.; Dalenberg, K.; Duarte, J.M.; Dutta, S.; et al. RCSB Protein Data Bank: Biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019, 47, D464–D474. [Google Scholar] [CrossRef] [Green Version]
- Artymiuk, P.J.; Green, J. The double life of aconitase. Structure 2006, 14, 2–4. [Google Scholar] [CrossRef]
- Banerjee, S.; Nandyala, A.K.; Raviprasad, P.; Ahmed, N.; Hasnain, S.E. Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J. Bacteriol. 2007, 189, 4046–4052. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, M.C.; Mende-Mueller, L.; Blondin, G.A.; Beinert, H. Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc. Natl. Acad. Sci. USA 1992, 89, 11730–11734. [Google Scholar] [CrossRef] [Green Version]
- Howard, P.K.; Shaw, J.; Otsuka, A.J. Nucleotide sequence of the birA gene encoding the biotin operon repressor and biotin holoenzyme synthetase functions of Escherichia coli. Gene 1985, 35, 321–331. [Google Scholar] [CrossRef]
- Barker, D.F.; Campbell, A.M. Genetic and biochemical characterization of the birA gene and its product: Evidence for a direct role of biotin holoenzyme synthetase in repression of the biotin operon in Escherichia coli. J. Mol. Biol. 1981, 146, 469–492. [Google Scholar] [CrossRef]
- Wan, F.; Anderson, D.E.; Barnitz, R.A.; Snow, A.; Bidere, N.; Zheng, L.; Hegde, V.; Lam, L.T.; Staudt, L.M.; Levens, D.; et al. Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell 2007, 131, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Malygin, A.A.; Parakhnevitch, N.M.; Ivanov, A.V.; Eperon, I.C.; Karpova, G.G. Human ribosomal protein S13 regulates expression of its own gene at the splicing step by a feedback mechanism. Nucleic Acids Res. 2007, 35, 6414–6423. [Google Scholar] [CrossRef] [Green Version]
- Fewell, S.W.; Woolford, J.L., Jr. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell Biol. 1999, 19, 826–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presutti, C.; Ciafre, S.A.; Bozzoni, I. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 1991, 10, 2215–2221. [Google Scholar] [CrossRef]
- Zengel, J.M.; Lindahl, L. A hairpin structure upstream of the terminator hairpin required for ribosomal protein L4-mediated attenuation control of the S10 operon of Escherichia coli. J. Bacteriol. 1996, 178, 2383–2387. [Google Scholar] [CrossRef] [Green Version]
- Horn, H.F.; Vousden, K.H. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 2008, 27, 5774–5784. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, J.F.; Tran, E.J.; Maxwell, E.S. Archaeal ribosomal protein L7 is a functional homolog of the eukaryotic 15.5kD/Snu13p snoRNP core protein. Nucleic Acids Res. 2002, 30, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Lohrum, M.A.; Ludwig, R.L.; Kubbutat, M.H.; Hanlon, M.; Vousden, K.H. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 2003, 3, 577–587. [Google Scholar] [CrossRef] [Green Version]
- Mazumder, B.; Sampath, P.; Seshadri, V.; Maitra, R.K.; DiCorleto, P.E.; Fox, P.L. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 2003, 115, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Dai, M.S.; Zeng, S.X.; Jin, Y.; Sun, X.X.; David, L.; Lu, H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell Biol. 2004, 24, 7654–7668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takagi, M.; Absalon, M.J.; McLure, K.G.; Kastan, M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 2005, 123, 49–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, D.M., 3rd; Deutsch, W.A.; Kelley, M.R. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 1994, 269, 25359–25364. [Google Scholar] [PubMed]
- Chen, X.J.; Wang, X.; Kaufman, B.A.; Butow, R.A. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 2005, 307, 714–717. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Krojer, T.; Garrido-Franco, M.; Huber, R.; Ehrmann, M.; Clausen, T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature 2002, 416, 455–459. [Google Scholar] [CrossRef]
- Jeffery, C.J. An enzyme in the test tube, and a transcription factor in the cell: Moonlighting proteins and cellular factors that affect their behavior. Protein Sci. 2019, 28, 1233–1238. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Dunker, A.K. Natively disordered proteins: Functions and predictions. Appl. Bioinform. 2004, 3, 105–113. [Google Scholar] [CrossRef]
- Dyson, H.J. Making Sense of Intrinsically Disordered Proteins. Biophys. J. 2016, 110, 1013–1016. [Google Scholar] [CrossRef] [Green Version]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef] [Green Version]
- Tompa, P.; Szasz, C.; Buday, L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005, 30, 484–489. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef] [Green Version]
- Hatos, A.; Hajdu-Soltesz, B.; Monzon, A.M.; Palopoli, N.; Alvarez, L.; Aykac-Fas, B.; Bassot, C.; Benitez, G.I.; Bevilacqua, M.; Chasapi, A.; et al. DisProt: Intrinsic protein disorder annotation in 2020. Nucleic Acids Res. 2020, 48, D269–D276. [Google Scholar] [CrossRef] [Green Version]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Green, D.R.; Kroemer, G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009, 458, 1127–1130. [Google Scholar] [CrossRef]
- Zhou, J.; Ahn, J.; Wilson, S.H.; Prives, C. A role for p53 in base excision repair. EMBO J. 2001, 20, 914–923. [Google Scholar] [CrossRef] [Green Version]
- Clore, G.M.; Omichinski, J.G.; Sakaguchi, K.; Zambrano, N.; Sakamoto, H.; Appella, E.; Gronenborn, A.M. High-resolution structure of the oligomerization domain of p53 by multidimensional NMR. Science 1994, 265, 386–391. [Google Scholar] [CrossRef]
- Wells, M.; Tidow, H.; Rutherford, T.J.; Markwick, P.; Jensen, M.R.; Mylonas, E.; Svergun, D.I.; Blackledge, M.; Fersht, A.R. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. USA 2008, 105, 5762–5767. [Google Scholar] [CrossRef] [Green Version]
- Bell, S.; Klein, C.; Muller, L.; Hansen, S.; Buchner, J. p53 contains large unstructured regions in its native state. J. Mol. Biol. 2002, 322, 917–927. [Google Scholar] [CrossRef]
- Safer, D.; Elzinga, M.; Nachmias, V.T. Thymosin beta 4 and Fx, an actin-sequestering peptide, are indistinguishable. J. Biol. Chem. 1991, 266, 4029–4032. [Google Scholar] [PubMed]
- Cassimeris, L.; Safer, D.; Nachmias, V.T.; Zigmond, S.H. Thymosin beta 4 sequesters the majority of G-actin in resting human polymorphonuclear leukocytes. J. Cell Biol. 1992, 119, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.S.; Rose, W.; Yaen, C.; Goldstein, A.; Martinez, J.; Kleinman, H. Thymosin beta4 enhances endothelial cell differentiation and angiogenesis. Angiogenesis 1999, 3, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Smart, N.; Rossdeutsch, A.; Riley, P.R. Thymosin beta4 and angiogenesis: Modes of action and therapeutic potential. Angiogenesis 2007, 10, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Philp, D.; Kleinman, H.K. Animal studies with thymosin beta, a multifunctional tissue repair and regeneration peptide. Ann. NY Acad. Sci. 2010, 1194, 81–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, J.D.; Lawrence, A.J.; MacLean, A.G.; Leung, B.P.; McInnes, I.B.; Canas, B.; Pappin, D.J.; Stevenson, R.D. Thymosin beta 4 sulfoxide is an anti-inflammatory agent generated by monocytes in the presence of glucocorticoids. Nat. Med. 1999, 5, 1424–1427. [Google Scholar] [CrossRef] [PubMed]
- Safer, D.; Sosnick, T.R.; Elzinga, M. Thymosin beta 4 binds actin in an extended conformation and contacts both the barbed and pointed ends. Biochemistry 1997, 36, 5806–5816. [Google Scholar] [CrossRef]
- Domanski, M.; Hertzog, M.; Coutant, J.; Gutsche-Perelroizen, I.; Bontems, F.; Carlier, M.F.; Guittet, E.; van Heijenoort, C. Coupling of folding and binding of thymosin beta4 upon interaction with monomeric actin monitored by nuclear magnetic resonance. J. Biol. Chem. 2004, 279, 23637–23645. [Google Scholar] [CrossRef] [Green Version]
- Bock-Marquette, I.; Saxena, A.; White, M.D.; Dimaio, J.M.; Srivastava, D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004, 432, 466–472. [Google Scholar] [CrossRef]
- Ho, J.H.; Tseng, K.C.; Ma, W.H.; Chen, K.H.; Lee, O.K.; Su, Y. Thymosin beta-4 upregulates anti-oxidative enzymes and protects human cornea epithelial cells against oxidative damage. Br. J. Ophthalmol. 2008, 92, 992–997. [Google Scholar] [CrossRef] [PubMed]
- Tantos, A.; Szabo, B.; Lang, A.; Varga, Z.; Tsylonok, M.; Bokor, M.; Verebelyi, T.; Kamasa, P.; Tompa, K.; Perczel, A.; et al. Multiple fuzzy interactions in the moonlighting function of thymosin-beta4. Intrinsically Disord Proteins 2013, 1, e26204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Czisch, M.; Schleicher, M.; Horger, S.; Voelter, W.; Holak, T.A. Conformation of thymosin beta 4 in water determined by NMR spectroscopy. Eur. J. Biochem. 1993, 218, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Hope, I.A.; Struhl, K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 1986, 46, 885–894. [Google Scholar] [CrossRef]
- Nikolaev, Y.; Deillon, C.; Hoffmann, S.R.; Bigler, L.; Friess, S.; Zenobi, R.; Pervushin, K.; Hunziker, P.; Gutte, B. The leucine zipper domains of the transcription factors GCN4 and c-Jun have ribonuclease activity. PLoS ONE 2010, 5, e10765. [Google Scholar] [CrossRef]
- Weiss, M.A.; Ellenberger, T.; Wobbe, C.R.; Lee, J.P.; Harrison, S.C.; Struhl, K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature 1990, 347, 575–578. [Google Scholar] [CrossRef]
- Leon-Del-Rio, A.; Valadez-Graham, V.; Gravel, R.A. Holocarboxylase Synthetase: A Moonlighting Transcriptional Coregulator of Gene Expression and a Cytosolic Regulator of Biotin Utilization. Annu. Rev. Nutr. 2017, 37, 207–223. [Google Scholar] [CrossRef]
- Wilson, K.P.; Shewchuk, L.M.; Brennan, R.G.; Otsuka, A.J.; Matthews, B.W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. USA 1992, 89, 9257–9261. [Google Scholar] [CrossRef] [Green Version]
- Merenmies, J.; Pihlaskari, R.; Laitinen, J.; Wartiovaara, J.; Rauvala, H. 30-kDa heparin-binding protein of brain (amphoterin) involved in neurite outgrowth. Amino acid sequence and localization in the filopodia of the advancing plasma membrane. J. Biol. Chem. 1991, 266, 16722–16729. [Google Scholar]
- Hardman, C.H.; Broadhurst, R.W.; Raine, A.R.; Grasser, K.D.; Thomas, J.O.; Laue, E.D. Structure of the A-domain of HMG1 and its interaction with DNA as studied by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry 1995, 34, 16596–16607. [Google Scholar] [CrossRef]
- Stott, K.; Watson, M.; Howe, F.S.; Grossmann, J.G.; Thomas, J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J. Mol. Biol. 2010, 403, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Brunagel, G.; Shah, U.; Schoen, R.E.; Getzenberg, R.H. Identification of calreticulin as a nuclear matrix protein associated with human colon cancer. J. Cell Biochem. 2003, 89, 238–243. [Google Scholar] [CrossRef] [PubMed]
- Goicoechea, S.; Orr, A.W.; Pallero, M.A.; Eggleton, P.; Murphy-Ullrich, J.E. Thrombospondin mediates focal adhesion disassembly through interactions with cell surface calreticulin. J. Biol. Chem. 2000, 275, 36358–36368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvier, M.; Stafford, W.F. Probing the three-dimensional structure of human calreticulin. Biochemistry 2000, 39, 14950–14959. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.L.; Nomura, M.E. coli ribosomal protein L4 is a feedback regulatory protein. Cell 1980, 21, 517–522. [Google Scholar] [CrossRef]
- Timsit, Y.; Acosta, Z.; Allemand, F.; Chiaruttini, C.; Springer, M. The role of disordered ribosomal protein extensions in the early steps of eubacterial 50 S ribosomal subunit assembly. Int. J. Mol. Sci. 2009, 10, 817–834. [Google Scholar] [CrossRef] [Green Version]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef]
- Varma, D.; Chandrasekaran, S.; Sundin, L.J.; Reidy, K.T.; Wan, X.; Chasse, D.A.; Nevis, K.R.; DeLuca, J.G.; Salmon, E.D.; Cook, J.G. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat. Cell Biol. 2012, 14, 593–603. [Google Scholar] [CrossRef] [Green Version]
- Khayrutdinov, B.I.; Bae, W.J.; Yun, Y.M.; Lee, J.H.; Tsuyama, T.; Kim, J.J.; Hwang, E.; Ryu, K.S.; Cheong, H.K.; Cheong, C.; et al. Structure of the Cdt1 C-terminal domain: Conservation of the winged helix fold in replication licensing factors. Protein Sci. 2009, 18, 2252–2264. [Google Scholar] [CrossRef] [Green Version]
- Coschigano, P.W.; Magasanik, B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol. Cell Biol. 1991, 11, 822–832. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.; Zhou, J.M.; Perrett, S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J. Biol. Chem. 2004, 279, 50025–50030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierce, M.M.; Baxa, U.; Steven, A.C.; Bax, A.; Wickner, R.B. Is the prion domain of soluble Ure2p unstructured? Biochemistry 2005, 44, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Costi, M.P.; Ferrari, S.; Venturelli, A.; Calo, S.; Tondi, D.; Barlocco, D. Thymidylate synthase structure, function and implication in drug discovery. Curr. Med. Chem. 2005, 12, 2241–2258. [Google Scholar] [CrossRef] [PubMed]
- Chu, E.; Koeller, D.M.; Casey, J.L.; Drake, J.C.; Chabner, B.A.; Elwood, P.C.; Zinn, S.; Allegra, C.J. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. USA 1991, 88, 8977–8981. [Google Scholar] [CrossRef] [Green Version]
- Phan, J.; Steadman, D.J.; Koli, S.; Ding, W.C.; Minor, W.; Dunlap, R.B.; Berger, S.H.; Lebioda, L. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 2001, 276, 14170–14177. [Google Scholar] [CrossRef] [Green Version]
- Margoliash, E.; Smith, E.L.; Kreil, G.; Tuppy, H. Amino-acid sequence of horse heart cytochrome c. Nature 1961, 192, 1125–1127. [Google Scholar] [CrossRef]
- Yu, T.; Wang, X.; Purring-Koch, C.; Wei, Y.; McLendon, G.L. A mutational epitope for cytochrome C binding to the apoptosis protease activation factor-1. J. Biol. Chem. 2001, 276, 13034–13038. [Google Scholar] [CrossRef] [Green Version]
- Stellwagen, E.; Rysavy, R.; Babul, G. The conformation of horse heart apocytochrome c. J. Biol. Chem. 1972, 247, 8074–8077. [Google Scholar]
- Lux, S.E.; John, K.M.; Kopito, R.R.; Lodish, H.F. Cloning and characterization of band 3, the human erythrocyte anion-exchange protein (AE1). Proc. Natl. Acad. Sci. USA 1989, 86, 9089–9093. [Google Scholar] [CrossRef] [Green Version]
- Low, P.S.; Rathinavelu, P.; Harrison, M.L. Regulation of glycolysis via reversible enzyme binding to the membrane protein, band 3. J. Biol. Chem. 1993, 268, 14627–14631. [Google Scholar]
- Zhang, D.; Kiyatkin, A.; Bolin, J.T.; Low, P.S. Crystallographic structure and functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. Blood 2000, 96, 2925–2933. [Google Scholar] [CrossRef]
- Zielenski, J.; Rozmahel, R.; Bozon, D.; Kerem, B.; Grzelczak, Z.; Riordan, J.R.; Rommens, J.; Tsui, L.C. Genomic DNA sequence of the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Genomics 1991, 10, 214–228. [Google Scholar] [CrossRef]
- Stutts, M.J.; Canessa, C.M.; Olsen, J.C.; Hamrick, M.; Cohn, J.A.; Rossier, B.C.; Boucher, R.C. CFTR as a cAMP-dependent regulator of sodium channels. Science 1995, 269, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Ostedgaard, L.S.; Baldursson, O.; Vermeer, D.W.; Welsh, M.J.; Robertson, A.D. A functional R domain from cystic fibrosis transmembrane conductance regulator is predominantly unstructured in solution. Proc. Natl. Acad. Sci. USA 2000, 97, 5657–5662. [Google Scholar] [CrossRef] [Green Version]
- Fuxreiter, M. Fuzziness in Protein Interactions-A Historical Perspective. J. Mol. Biol. 2018, 430, 2278–2287. [Google Scholar] [CrossRef]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: Polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008, 33, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Raduly, Z.; Miskei, M.; Fuxreiter, M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015, 589, 2533–2542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uversky, V.N. Supramolecular Fuzziness of Intracellular Liquid Droplets: Liquid-Liquid Phase Transitions, Membrane-Less Organelles, and Intrinsic Disorder. Molecules 2019, 24, 3265. [Google Scholar] [CrossRef] [Green Version]
- Miskei, M.; Antal, C.; Fuxreiter, M. FuzDB: Database of fuzzy complexes, a tool to develop stochastic structure-function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res. 2017, 45, D228–D235. [Google Scholar] [CrossRef] [Green Version]
- Murzin, A.G. Biochemistry. Metamorphic proteins. Science 2008, 320, 1725–1726. [Google Scholar] [CrossRef]
- Dishman, A.F.; Volkman, B.F. Unfolding the Mysteries of Protein Metamorphosis. Acs. Chem. Biol. 2018, 13, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Lella, M.; Mahalakshmi, R. Metamorphic Proteins: Emergence of Dual Protein Folds from One Primary Sequence. Biochemistry 2017, 56, 2971–2984. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, S.C.; Curmi, P.M.G.; Brown, L.J. Structural gymnastics of multifunctional metamorphic proteins. Biophys. Rev. 2011, 3, 143. [Google Scholar] [CrossRef] [PubMed]
- Tuinstra, R.L.; Peterson, F.C.; Kutlesa, S.; Elgin, E.S.; Kron, M.A.; Volkman, B.F. Interconversion between two unrelated protein folds in the lymphotactin native state. Proc. Natl. Acad. Sci. USA 2008, 105, 5057–5062. [Google Scholar] [CrossRef] [Green Version]
- Burmann, B.M.; Knauer, S.H.; Sevostyanova, A.; Schweimer, K.; Mooney, R.A.; Landick, R.; Artsimovitch, I.; Rosch, P. An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 2012, 150, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Goodchild, S.C.; Howell, M.W.; Littler, D.R.; Mandyam, R.A.; Sale, K.L.; Mazzanti, M.; Breit, S.N.; Curmi, P.M.; Brown, L.J. Metamorphic response of the CLIC1 chloride intracellular ion channel protein upon membrane interaction. Biochemistry 2010, 49, 5278–5289. [Google Scholar] [CrossRef]
- Luo, X.; Yu, H. Protein metamorphosis: The two-state behavior of Mad2. Structure 2008, 16, 1616–1625. [Google Scholar] [CrossRef] [Green Version]
- Tseng, R.; Goularte, N.F.; Chavan, A.; Luu, J.; Cohen, S.E.; Chang, Y.G.; Heisler, J.; Li, S.; Michael, A.K.; Tripathi, S.; et al. Structural basis of the day-night transition in a bacterial circadian clock. Science 2017, 355, 1174–1180. [Google Scholar] [CrossRef] [Green Version]
- Dai, Z.; Tonelli, M.; Markley, J.L. Metamorphic protein IscU changes conformation by cis-trans isomerizations of two peptidyl-prolyl peptide bonds. Biochemistry 2012, 51, 9595–9602. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Pelegrin, M.; Cerda-Costa, N.; Cintas-Pedrola, A.; Herranz-Trillo, F.; Bernado, P.; Peinado, J.R.; Arolas, J.L.; Gomis-Ruth, F.X. Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase. Angew. Chem. Int. Ed. Engl. 2014, 53, 10624–10630. [Google Scholar] [CrossRef] [Green Version]
- London, R.E. HIV-1 Reverse Transcriptase: A Metamorphic Protein with Three Stable States. Structure 2019, 27, 420–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peterson, F.C.; Elgin, E.S.; Nelson, T.J.; Zhang, F.; Hoeger, T.J.; Linhardt, R.J.; Volkman, B.F. Identification and characterization of a glycosaminoglycan recognition element of the C chemokine lymphotactin. J. Biol. Chem. 2004, 279, 12598–12604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuinstra, R.L.; Peterson, F.C.; Elgin, E.S.; Pelzek, A.J.; Volkman, B.F. An engineered second disulfide bond restricts lymphotactin/XCL1 to a chemokine-like conformation with XCR1 agonist activity. Biochemistry 2007, 46, 2564–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knauer, S.H.; Artsimovitch, I.; Rosch, P. Transformer proteins. Cell Cycle 2012, 11, 4289–4290. [Google Scholar] [CrossRef] [Green Version]
- Al Khamici, H.; Brown, L.J.; Hossain, K.R.; Hudson, A.L.; Sinclair-Burton, A.A.; Ng, J.P.; Daniel, E.L.; Hare, J.E.; Cornell, B.A.; Curmi, P.M.; et al. Members of the chloride intracellular ion channel protein family demonstrate glutaredoxin-like enzymatic activity. PLoS ONE 2015, 10, e115699. [Google Scholar] [CrossRef] [Green Version]
- Littler, D.R.; Harrop, S.J.; Fairlie, W.D.; Brown, L.J.; Pankhurst, G.J.; Pankhurst, S.; DeMaere, M.Z.; Campbell, T.J.; Bauskin, A.R.; Tonini, R.; et al. The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J. Biol. Chem. 2004, 279, 9298–9305. [Google Scholar] [CrossRef] [Green Version]
- Warton, K.; Tonini, R.; Fairlie, W.D.; Matthews, J.M.; Valenzuela, S.M.; Qiu, M.R.; Wu, W.M.; Pankhurst, S.; Bauskin, A.R.; Harrop, S.J.; et al. Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J. Biol. Chem. 2002, 277, 26003–26011. [Google Scholar] [CrossRef] [Green Version]
- Jaffe, E.K. Morpheeins—A new structural paradigm for allosteric regulation. Trends Biochem. Sci. 2005, 30, 490–497. [Google Scholar] [CrossRef]
- Jaffe, E.K.; Lawrence, S.H. Allostery and the dynamic oligomerization of porphobilinogen synthase. Arch. Biochem. Biophys. 2012, 519, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.G.; Gu, M.; Etlinger, J.D. 240-kDa proteasome inhibitor (CF-2) is identical to delta-aminolevulinic acid dehydratase. J. Biol. Chem. 1994, 269, 12399–12402. [Google Scholar]
- Li, X.C.; Gu, M.Z.; Etlinger, J.D. Isolation and characterization of a novel endogenous inhibitor of the proteasome. Biochemistry 1991, 30, 9709–9715. [Google Scholar] [CrossRef] [PubMed]
- Breinig, S.; Kervinen, J.; Stith, L.; Wasson, A.S.; Fairman, R.; Wlodawer, A.; Zdanov, A.; Jaffe, E.K. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nat. Struct. Biol. 2003, 10, 757–763. [Google Scholar] [CrossRef] [PubMed]
- Kervinen, J.; Jaffe, E.K.; Stauffer, F.; Neier, R.; Wlodawer, A.; Zdanov, A. Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity. Biochemistry 2001, 40, 8227–8236. [Google Scholar] [CrossRef] [Green Version]
- Bornholdt, Z.A.; Noda, T.; Abelson, D.M.; Halfmann, P.; Wood, M.R.; Kawaoka, Y.; Saphire, E.O. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013, 154, 763–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, J.; Poruri, K.; Boniecki, M.T.; McTavish, K.K.; Martinis, S.A. Yeast mitochondrial leucyl-tRNA synthetase CP1 domain has functionally diverged to accommodate RNA splicing at expense of hydrolytic editing. J. Biol. Chem. 2012, 287, 14772–14781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef] [Green Version]
- Eyers, P.A.; Murphy, J.M. The evolving world of pseudoenzymes: Proteins, prejudice and zombies. Bmc Biol. 2016, 14, 98. [Google Scholar] [CrossRef] [Green Version]
- Murphy, J.M.; Farhan, H.; Eyers, P.A. Bio-Zombie: The rise of pseudoenzymes in biology. Biochem. Soc. Trans. 2017, 45, 537–544. [Google Scholar] [CrossRef]
- Jeffery, C.J. The demise of catalysis, but new functions arise: Pseudoenzymes as the phoenixes of the protein world. Biochem. Soc. Trans. 2019, 47, 371–379. [Google Scholar] [CrossRef]
- Todd, A.E.; Orengo, C.A.; Thornton, J.M. Sequence and structural differences between enzyme and nonenzyme homologs. Structure 2002, 10, 1435–1451. [Google Scholar] [CrossRef] [Green Version]
- Pils, B.; Schultz, J. Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 2004, 340, 399–404. [Google Scholar] [CrossRef]
- Qasba, P.K.; Safaya, S.K. Similarity of the nucleotide sequences of rat alpha-lactalbumin and chicken lysozyme genes. Nature 1984, 308, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Brew, K.; Vanaman, T.C.; Hill, R.L. Comparison of the amino acid sequence of bovine alpha-lactalbumin and hens egg white lysozyme. J. Biol. Chem. 1967, 242, 3747–3749. [Google Scholar] [PubMed]
- Powell, J.T.; Brew, K. On the interaction of alpha-lactalbumin and galactosyltransferase during lactose synthesis. J. Biol. Chem. 1975, 250, 6337–6343. [Google Scholar] [PubMed]
- Piatigorsky, J.; O’Brien, W.E.; Norman, B.L.; Kalumuck, K.; Wistow, G.J.; Borras, T.; Nickerson, J.M.; Wawrousek, E.F. Gene sharing by delta-crystallin and argininosuccinate lyase. Proc. Natl. Acad. Sci. USA 1988, 85, 3479–3483. [Google Scholar] [CrossRef] [Green Version]
- Sampaleanu, L.M.; Vallee, F.; Slingsby, C.; Howell, P.L. Structural studies of duck delta 1 and delta 2 crystallin suggest conformational changes occur during catalysis. Biochemistry 2001, 40, 2732–2742. [Google Scholar] [CrossRef]




| Protein | Organism | Function 1 | Function 1 Location | Function 2 | Function 2 Location | Disorder Content | UniProt ID | Ref. |
| P53 | Homo sapiens | Binds to regulatory element in the genome | Nucleus | Centrosome duplication, induction of apoptosis, autophagy inhibition | Cytoplasm | 37.00% | P04637 | [67,69,71] |
| Thymosin beta-4 | Homo sapiens | Involved in sequestering G-actin in human polymorphonuclear leukocytes | Cytoplasm | Secreted anti-inflammatory agent | Extracellular | 100.00% | P62328 | [73,78,84] |
| General control protein GCN4 | Saccharomyces cerevisiae | Transcription factor | Nucleus | Ribonuclease | Cytoplasm | 32.38% | P03069 | [85,86,87] |
| Bifunctional ligase/repressor BirA | Escherichia coli | Biotin synthetase, biotin–[acetyl–CoA-carboxylase] ligase | Cytoplasm | Biotin operon repressor, activity depends on cellular concentration of biotin | Bound to DNA | 7.17% | P06709 | [42,43,88,89] |
| High mobility group protein B1 | Rattus norvegicus | Binds heparin. | Cytoplasm | DNA binding protein, without sequence specificity | Nucleus | 35.81% | P63159 | [90,91,92] |
| Calreticulin | Homo sapiens | Protein-folding chaperone | Endoplasmic reticulum | Adhesin | Cell surface | 100.00% | P27797 | [93,94,95] |
| 50S ribosomal protein L4 | Escherichia coli | Ribosomal protein, part of the 50S subunit | Cytoplasm | Transcriptional repressor, causes premature termination of transcription | Bound to DNA | 31.34% | P60723 | [48,96,97] |
| DNA replication factor Cdt1 | Homo sapiens | Helps with initiating DNA replication. | Nucleus | Role in mitosis, localizes to kinetochores through binding to Ndc80 complex | Cytoplasm | 2.20% | Q9H211 | [98,99,100] |
| Transcriptional regulator Ure2 | Saccharomyce cerevisiae | Binds to and inhibits GATA transcriptional activators GLN3 and GAT1 | Cytoplasm | Glutathione peroxidase, thiol:disulfide oxidoreductase | Cytoplasm | 31.64% | P23202 | [101,102,103] |
| Thymidylate synthase | Homo sapiens | Thymidylate synthase | Cytoplasm | mRNA translation inhibition | Cytoplasm | 15.65% | P04818 | [104,105,106] |
| Cytochrome c | Equus caballus | Electron carrier protein | Mitochondrion | Binds to apoptosis protease activation factor-1 and promotes apoptosis | Cytoplasm | 99.05% | P00004 | [107,108,109] |
| Band 3 anion transport protein | Homo sapiens | Transports inorganic anions across the plasma membrane | Plasma membrane | Scaffold protein providing binding sites for glycolytic enzymes | Plasma membrane | 5.93% | P02730 | [110,111,112] |
| Cystic fibrosis transmembrane conductance regulator CFTR | Homo sapiens | Chloride transporter, contains nucleotide binding domains that bind and hydrolyze ATP | Plasma membrane | Regulator of other ion channels | Plasma membrane | 12.50% | P13569 | [113,114,115] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Jeffery, C.J. Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism. Molecules 2020, 25, 3440. https://doi.org/10.3390/molecules25153440
Liu H, Jeffery CJ. Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism. Molecules. 2020; 25(15):3440. https://doi.org/10.3390/molecules25153440
Chicago/Turabian StyleLiu, Haipeng, and Constance J. Jeffery. 2020. "Moonlighting Proteins in the Fuzzy Logic of Cellular Metabolism" Molecules 25, no. 15: 3440. https://doi.org/10.3390/molecules25153440