Discovery of New Inhibitors of eEF2K from Traditional Chinese Medicine Based on In Silico Screening and In Vitro Experimental Validation
Abstract
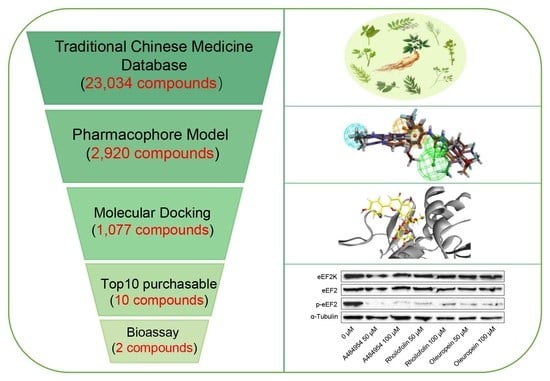
:1. Introduction
2. Results
2.1. Pharmacophore Model Generation and Validation
2.2. Virtual Screening Based on Pharmacophore Model
2.3. Homology Modeling
2.4. Virtual Screening Based on Molecular Docking
2.5. Characterization of the Binding of Rhoifolin and Oleuropein to eEF2K
2.6. Evaluation of Effects of Rhoifolin and Oleuropein on eEF2K Activity
2.7. Molecular Dynamics Simulation
2.8. Pharmacokinetic and Toxicological Analyses
3. Discussion
4. Materials and Methods
4.1. Construction of Pharmacophore Model
4.1.1. Preparation of the Training Set Molecules
4.1.2. Common Feature Pharmacophore Generation and Evaluation
4.1.3. Discovery of Lead Compounds
4.2. Homology Modeling
4.3. Virtual Screening Based on Molecular Docking
4.4. Chemicals
4.5. Cell Culture
4.6. Cell Viability Assay
4.7. Western Blotting
4.8. Molecular Dynamic Simulation
4.9. Pharmacokinetics and Toxicological Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1151–1210. [Google Scholar] [CrossRef] [Green Version]
- Ballard, D.J.; Peng, H.Y.; Das, J.K.; Kumar, A.; Wang, L.; Ren, Y.; Xiong, X.; Ren, X.; Yang, J.M.; Song, J. Insights into the Pathologic Roles and Regulation of Eukaryotic Elongation Factor-2 Kinase. Front. Mol. Biosci. 2021, 8, 727863. [Google Scholar] [CrossRef] [PubMed]
- Ryazanov, A.G.; Pavur, K.S.; Dorovkov, M.V. Alpha-kinases: A new class of protein kinases with a novel catalytic domain. Curr. Biol. 1999, 9, R43–R45. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, U.; Nilsson, A.; Nygård, O. Functional properties of phosphorylated elongation factor 2. Eur. J. Biochem. 1990, 191, 639–645. [Google Scholar] [CrossRef]
- Ryazanov, A.G.; Natapov, P.G.; Shestakova, E.A.; Severin, F.F.; Spirin, A.S. Phosphorylation of the elongation factor 2: The fifth Ca2+/calmodulin-dependent system of protein phosphorylation. Biochimie 1988, 70, 619–626. [Google Scholar] [CrossRef]
- Merrick, W.C.; Nyborg, J. The protein biosynthesis elongation cycle. Cold Spring Harb. Monogr. Ser. 2000, 39, 89–126. [Google Scholar]
- Zhu, H.; Yang, X.; Liu, J.; Zhou, L.; Zhang, C.; Xu, L.; Qin, Q.; Zhan, L.; Lu, J.; Cheng, H.; et al. Eukaryotic elongation factor 2 kinase confers tolerance to stress conditions in cancer cells. Cell Stress Chaperones 2015, 20, 217–220. [Google Scholar] [CrossRef] [Green Version]
- Leprivier, G.; Remke, M.; Rotblat, B.; Dubuc, A.; Mateo, A.R.; Kool, M.; Agnihotri, S.; El-Naggar, A.; Yu, B.; Somasekharan, S.P.; et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 2013, 153, 1064–1079. [Google Scholar] [CrossRef] [Green Version]
- Erdogan, M.A.; Ashour, A.; Yuca, E.; Gorgulu, K.; Ozpolat, B. Targeting eukaryotic elongation factor-2 kinase suppresses the growth and peritoneal metastasis of ovarian cancer. Cell Signal. 2021, 81, 109938. [Google Scholar] [CrossRef]
- Zhu, S.; Liao, M.; Tan, H.; Zhu, L.; Chen, Y.; He, G.; Liu, B. Inhibiting Eukaryotic Elongation Factor 2 Kinase: An Update on Pharmacological Small-Molecule Compounds in Cancer. J. Med. Chem. 2021, 64, 8870–8883. [Google Scholar] [CrossRef]
- Fu, L.L.; Xie, T.; Zhang, S.Y.; Liu, B. Eukaryotic elongation factor-2 kinase (eEF2K): A potential therapeutic target in cancer. Apoptosis 2014, 19, 1527–1531. [Google Scholar] [CrossRef] [PubMed]
- Karakas, D.; Ozpolat, B. Eukaryotic elongation factor-2 kinase (eEF2K) signaling in tumor and microenvironment as a novel molecular target. J. Mol. Med. 2020, 98, 775–787. [Google Scholar] [CrossRef] [PubMed]
- Temme, L.; Asquith, C.R.M. eEF2K: An atypical kinase target for cancer. Nat. Rev. Drug Discov. 2021, 20, 577. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Gopalakrishnan, S.M.; Bui, M.H.; Soni, N.B.; Warrior, U.; Johnson, E.F.; Donnelly, J.B.; Glaser, K.B. 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): A cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J. Biol. Chem. 2011, 286, 43951–43958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, T.; Liu, R.; Proud, C.G.; Wang, M.W. A high-throughput screening assay for eukaryotic elongation factor 2 kinase inhibitors. Acta Pharm. Sin. B 2016, 6, 557–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockman, J.W.; Reeder, M.D.; Suzuki, K.; Ostanin, K.; Hoff, R.; Bhoite, L.; Austin, H.; Baichwal, V.; Adam Willardsen, J. Inhibition of eEF2-K by thieno[2,3-b]pyridine analogues. Bioorg. Med. Chem. Lett. 2010, 20, 2283–2286. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, X.; Lin, X. A Review on Applications of Computational Methods in Drug Screening and Design. Molecules 2020, 25, 1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, W.-L.; Zhang, L.-X.; Guan, Y.-D.; Xue, W.-W.; Chen, A.F.; Cao, Q.; Cheng, Y.; Cao, D.-S. Virtual screening and experimental validation of eEF2K inhibitors by combining homology modeling, QSAR and molecular docking from FDA approved drugs. N. J. Chem. 2019, 43, 19097–19106. [Google Scholar] [CrossRef]
- Yoshimori, A.; Kawasaki, E.; Murakami, R.; Kanai, C. Discovery of Novel eEF2K Inhibitors Using HTS Fingerprint Generated from Predicted Profiling of Compound-Protein Interactions. Medicines 2021, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Bai, X.Y.; Wang, C.H. Traditional Chinese medicine: A treasured natural resource of anticancer drug research and development. Am. J. Chin. Med. 2014, 42, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hu, X.; Wang, S.; Jiao, Z.; Sun, T.; Liu, T.; Song, K. Characterization and anti-tumor bioactivity of astragalus polysaccharides by immunomodulation. Int. J. Biol. Macromol. 2020, 145, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.Y.; Han, Y.; Ahn, J.Y.; Yun, Y.S.; Song, J.Y. Chemoprotective and adjuvant effects of immunomodulator ginsan in cyclophosphamide-treated normal and tumor bearing mice. Int. J. Immunopathol. Pharmacol. 2007, 20, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zou, J.; Zhang, Q.; Wang, Z.; Wang, N.; He, S.; Zhao, Y.; Naman, C.B. Progress in the Development of Eukaryotic Elongation Factor 2 Kinase (eEF2K) Natural Product and Synthetic Small Molecule Inhibitors for Cancer Chemotherapy. Int. J. Mol. Sci. 2021, 22, 2408. [Google Scholar] [CrossRef] [PubMed]
- Comert Onder, F.; Durdagi, S.; Sahin, K.; Ozpolat, B.; Ay, M. Design, synthesis, and molecular modeling studies of novel coumarin carboxamide derivatives as eEF-2K inhibitors. J. Chem. Inf. Model. 2020, 60, 1766–1778. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, Y.; Wang, G.; Chen, Y.; Jiang, Y.; Ouyang, L.; Liu, B. Design, synthesis and structure-activity relationship of a focused library of β-phenylalanine derivatives as novel eEF2K inhibitors with apoptosis-inducing mechanisms in breast cancer. Eur. J. Med. Chem. 2018, 143, 402–418. [Google Scholar] [CrossRef]
- Pan, Z.; Chen, Y.; Liu, J.; Jiang, Q.; Yang, S.; Guo, L.; He, G. Design, synthesis, and biological evaluation of polo-like kinase 1/eukaryotic elongation factor 2 kinase (PLK1/EEF2K) dual inhibitors for regulating breast cancer cells apoptosis and autophagy. Eur. J. Med. Chem. 2018, 144, 517–528. [Google Scholar] [CrossRef]
- Piserchio, A.; Will, N.; Giles, D.H.; Hajredini, F.; Dalby, K.N.; Ghose, R. Solution structure of the carboxy-terminal tandem repeat domain of eukaryotic elongation factor 2 kinase and its role in substrate recognition. J. Mol. Biol. 2019, 431, 2700–2717. [Google Scholar] [CrossRef]
- Lin, J.H.; Yamazaki, M. Role of P-glycoprotein in pharmacokinetics: Clinical implications. Clin. Pharmacokinet. 2003, 42, 59–98. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.W.; Han, Y.Y.; Jia, X.S. PAFR-deficiency alleviates myocardial ischemia/reperfusion injury in mice via suppressing inflammation, oxidative stress and apoptosis. Biochem. Biophys. Res. Commun. 2018, 495, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Proud, C.G. Eukaryotic elongation factor 2 kinase as a drug target in cancer, and in cardiovascular and neurodegenerative diseases. Acta Pharmacol. Sin. 2016, 37, 285–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BIOVIA Discovery Studio. Available online: https://www.discngine.com/discovery-studio/ (accessed on 7 July 2022).
- Earl, L.A.; Falconieri, V.; Milne, J.L.; Subramaniam, S. Cryo-EM: Beyond the microscope. Curr. Opin. Struct. Biol. 2017, 46, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Hattori, S.; Matsuda, H. Rhoifolin, a new flavone glycoside, isolated from the leaves of Rhus succedanea. Arch. Biochem. Biophys. 1952, 37, 85–89. [Google Scholar] [CrossRef]
- Cárdeno, A.; Sánchez-Hidalgo, M.; Rosillo, M.A.; de la Lastra, C.A. Oleuropein, a secoiridoid derived from olive tree, inhibits the proliferation of human colorectal cancer cell through downregulation of HIF-1α. Nutr. Cancer 2013, 65, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Eldahshan, O.A. Rhoifolin; a potent antiproliferative effect on cancer cell lines. Br. J. Pharm. Res. 2013, 3, 46–53. [Google Scholar] [CrossRef]
- Shamshoum, H.; Vlavcheski, F.; Tsiani, E. Anticancer effects of oleuropein. Biofactors 2017, 43, 517–528. [Google Scholar] [CrossRef]
- Yoshida, T.; Miyazawa, K.; Kasuga, I.; Yokoyama, T.; Minemura, K.; Ustumi, K.; Aoshima, M.; Ohyashiki, K. Apoptosis induction of vitamin K2 in lung carcinoma cell lines: The possibility of vitamin K2 therapy for lung cancer. Int. J. Oncol. 2003, 23, 627–632. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Luo, G.; Zhang, X.; Lu, F.; Qiao, L.; He, W.; Li, G.; Zhang, Y. Discovery of Potential Inhibitors of Squalene Synthase from Traditional Chinese Medicine Based on Virtual Screening and In Vitro Evaluation of Lipid-Lowering Effect. Molecules 2018, 23, 1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ren, Z.; He, Y.; Xiang, Y.; Zhang, Y.; Qiao, Y. A combination of pharmacophore modeling, molecular docking and virtual screening for iNOS inhibitors from Chinese herbs. Biomed. Mater. Eng. 2014, 24, 1315–1322. [Google Scholar] [CrossRef]
- Wang, C.; Tang, K.; Dai, Y.; Jia, H.; Li, Y.; Gao, Z.; Wu, B. Identification, Characterization, and Site-Specific Mutagenesis of a Thermostable ω-Transaminase from Chloroflexi bacterium. ACS Omega 2021, 6, 17058–17070. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, Z.; Dou, G.; Lv, X.; Ge, J.; Wang, H.; Xie, H.; Zhu, D. Novel natural inhibitors targeting B-RAF(V600E) by computational study. Bioengineered 2021, 12, 2970–2983. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, C.; Svensson, F.; Zoufir, A.; Bender, A. eMolTox: Prediction of molecular toxicity with confidence. Bioinformatics 2018, 34, 2508–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Pharmacophore | Features | Rank | D | A | Ha | Ht | HRA | IEI | CAI |
|---|---|---|---|---|---|---|---|---|---|
| 08 | HHA | 188.548 | 19 | 13 | 12 | 18 | 92.31% | 0.974359 | 0.899408 |
| 02 | RHA | 206.975 | 19 | 13 | 9 | 14 | 69.23% | 0.93956 | 0.650465 |
| 06 | RHA | 200.597 | 19 | 13 | 9 | 14 | 69.23% | 0.93956 | 0.650465 |
| 01 | RHA | 209.374 | 19 | 13 | 8 | 13 | 61.54% | 0.899408 | 0.553482 |
| 09 | RHA | 187.640 | 19 | 13 | 5 | 8 | 38.46% | 0.913462 | 0.351331 |
| 03 | RHA | 206.677 | 19 | 13 | 2 | 3 | 15.38% | 0.974359 | 0.149901 |
| 05 | RHA | 201.811 | 19 | 13 | 2 | 4 | 15.38% | 0.730769 | 0.112426 |
| 07 | RHA | 191.351 | 19 | 13 | 2 | 2 | 15.38% | 1.461538 | 0.224852 |
| 04 | RHA | 201.811 | 19 | 13 | 0 | 0 | 0.00% | 0 | 0 |
| 10 | RHA | 185.059 | 19 | 13 | 0 | 0 | 0.00% | 0 | 0 |
| Name | PDF Total Energy | PDF Physical Energy | DOPE Score |
|---|---|---|---|
| M0007 | 18,709.2754 | 2533.81030749 | −20,998.439453 |
| M0003 | 19,247.8340 | 2847.16375490001 | −18,696.406250 |
| M0004 | 19,468.7598 | 2588.140841099 | −18,796.500000 |
| M0006 | 19,751.0020 | 2865.9443023 | −21,234.820312 |
| M0008 | 20,003.1426 | 2772.2228905791 | −22,023.769531 |
| M0005 | 20,003.2539 | 2956.111978 | −20,604.794922 |
| M0009 | 20,415.1406 | 2995.0741653 | −21,289.044922 |
| M0002 | 20,913.9961 | 3115.7153669 | −19,221.875000 |
| M0001 | 21,081.1328 | 3155.63869406 | −19,377.949219 |
| M0010 | 22,975.1074 | 3739.238885 | −19,577.082031 |
| Rank | Name | Source Plant | LibDock Score | Price |
|---|---|---|---|---|
| 1 | Oleuropein | Fraxinus chinensis, Ligustrum lucidum, Fraxinus japonica, Ligustrum japonicum, Olea europaea. | 152.589 | 1.80 $/mg |
| 2 | Rhoifolin | Anabasis aphylla | 153.939 | 5.85 $/mg |
| 3 | Vitamin K2 | Hippophae rhamnoides. | 152.964 | 39.46 $/mg |
| 4 | Licuroside | Glycyrrhiza sp | 148.674 | 63.25 $/mg |
| 5 | Chrysophanol-8-O-β-d-(6′-O-galloyl)-glucopyranoside | Rheum hotaoense. | 152.437 | 74.96 $/mg |
| 6 | Calyxin H | Alpinia pinnanensis | 160.676 | 90.36 $/mg |
| 7 | Sanggenon G | Morus mongolica, Morus alba. | 149.677 | 134.93 $/mg |
| 8 | Cannabisin D | Hyoscyamus niger | 154.383 | 154.33 $/mg |
| 9 | Bis-5,5-nortrachelogenin | Wikstroemia indica | 166.105 | 426.95 $/mg |
| 10 | Fortunellin | Fortunella margarita, Fortunella crassifolia | 160.992 | 619.72 $/mg |
| Pharmacokinetic Analyses | Oleuropein | Rhoifolin |
|---|---|---|
| GI absorption | Low | Low |
| BBB permeant | No | No |
| P-gp substrate | Yes | Yes |
| CYP1A2 inhibitor | No | No |
| CYP2C19 inhibitor | No | No |
| CYP2C9 inhibitor | No | No |
| CYP2D6 inhibitor | No | No |
| CYP3A4 inhibitor | No | No |
| Toxicological Analyses | Oleuropein | Rhoifolin |
|---|---|---|
| Cardiotoxicity | Negative | Positive, modulator of platelet activating factor receptor |
| CNS Toxicity | Negative | Negative |
| Mutagenicity Genotoxicity | Negative | Negative |
| Carcinogenicity | Negative | Negative |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Q.; Liu, X.; Li, Y.; Wang, P.; Wu, T.; Xiao, H.; Zhao, Y.; Liao, Q.; Song, Z. Discovery of New Inhibitors of eEF2K from Traditional Chinese Medicine Based on In Silico Screening and In Vitro Experimental Validation. Molecules 2022, 27, 4886. https://doi.org/10.3390/molecules27154886
Fu Q, Liu X, Li Y, Wang P, Wu T, Xiao H, Zhao Y, Liao Q, Song Z. Discovery of New Inhibitors of eEF2K from Traditional Chinese Medicine Based on In Silico Screening and In Vitro Experimental Validation. Molecules. 2022; 27(15):4886. https://doi.org/10.3390/molecules27154886
Chicago/Turabian StyleFu, Qinghua, Xiaomei Liu, Yan Li, Peng Wang, Tian Wu, Haihan Xiao, Yameng Zhao, Qichao Liao, and Ziyi Song. 2022. "Discovery of New Inhibitors of eEF2K from Traditional Chinese Medicine Based on In Silico Screening and In Vitro Experimental Validation" Molecules 27, no. 15: 4886. https://doi.org/10.3390/molecules27154886