Role of Helicity on the Anticancer Mechanism of Action of Cationic-Helical Peptides
Abstract
:1. Introduction
2. Results
2.1. Peptide Design
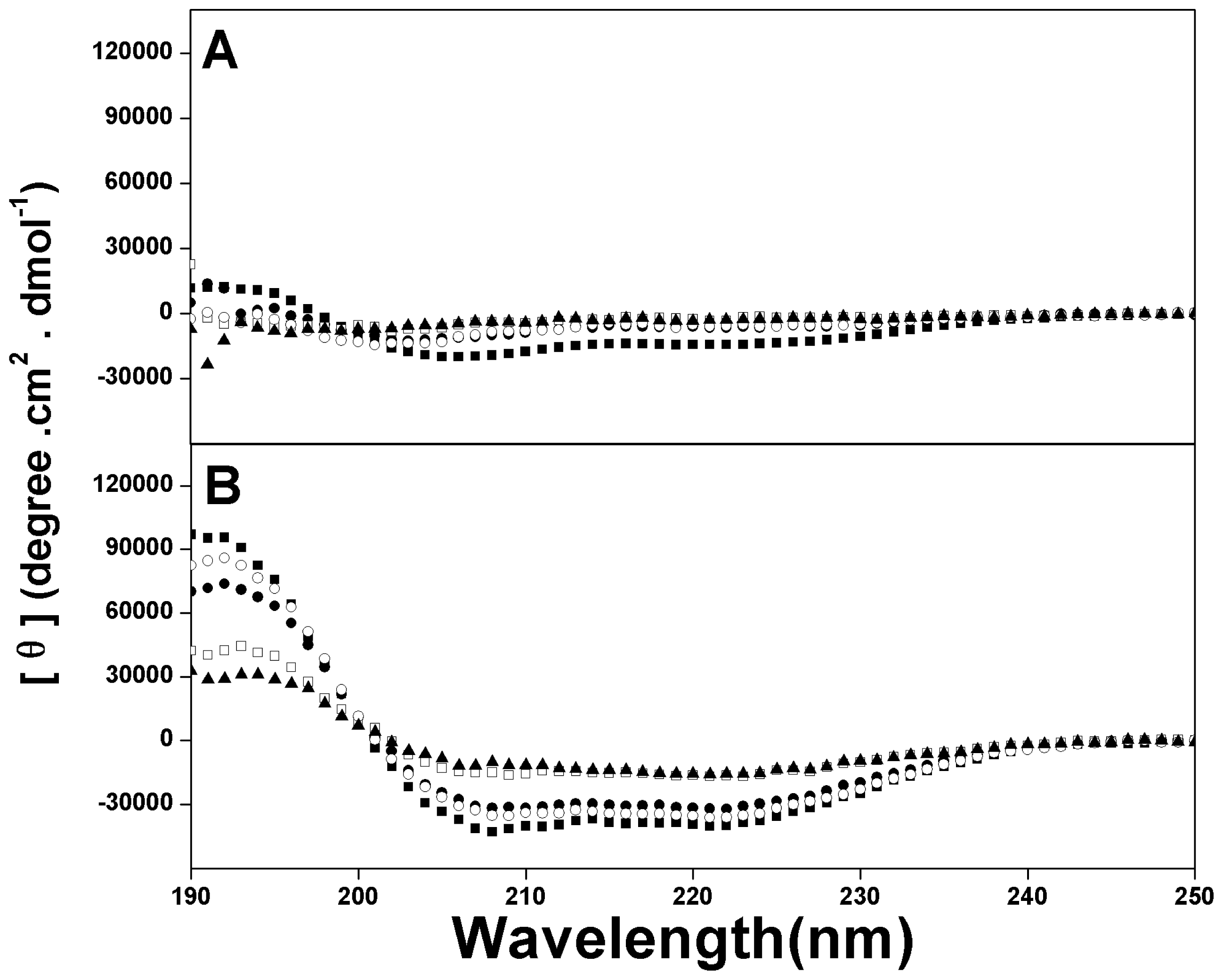
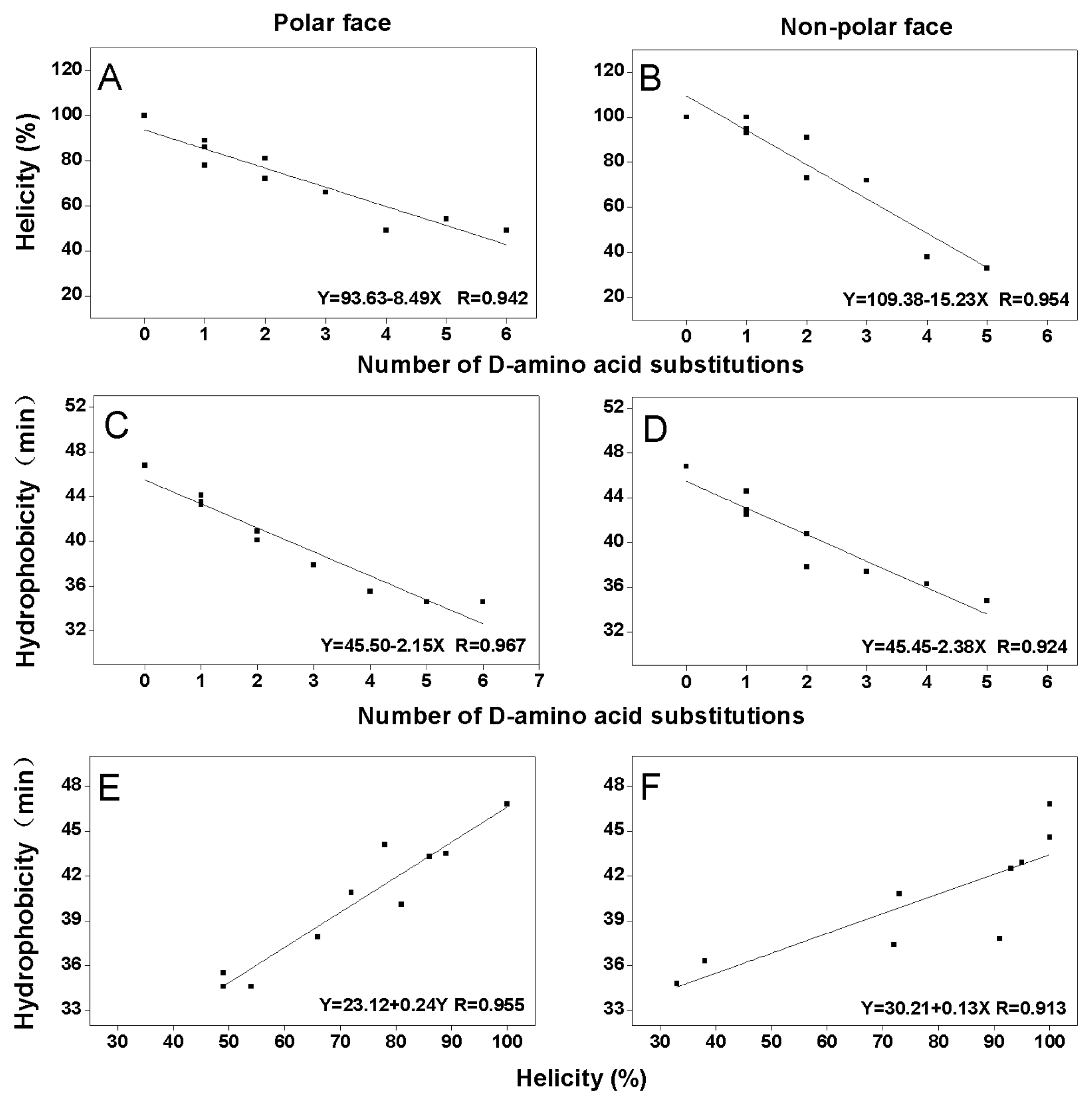
2.2. Peptide Secondary Structure
2.3. Peptide Hydrophobicity
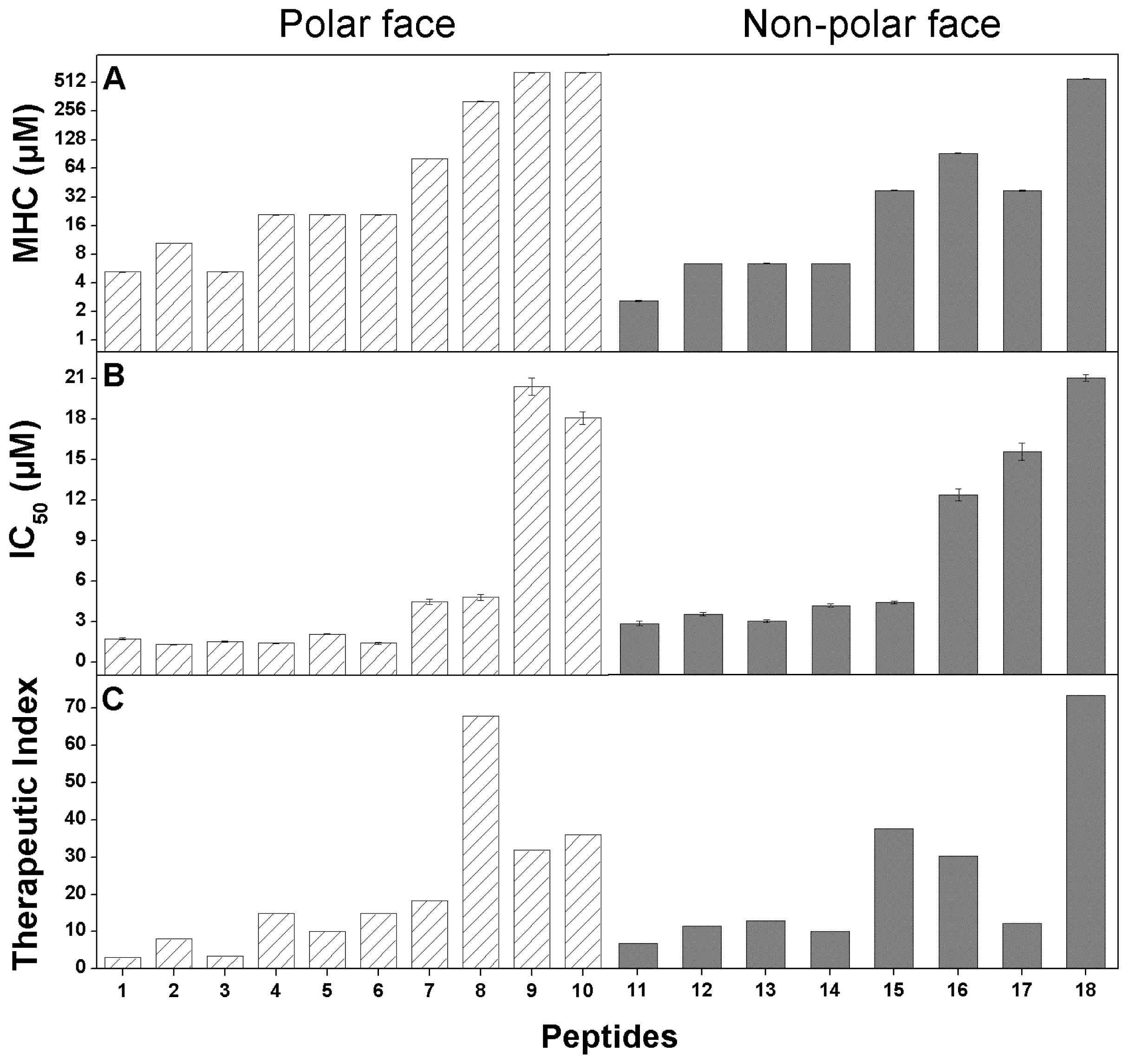
2.4. Anticancer Activity
2.5. Hemolytic Activity
2.6. Peptide Specificity (Therapeutic Index)
3. Discussion
4. Experimental Section
4.1. Materials
4.2. Cell Line and Cell Culture
4.3. Peptide Synthesis and Purification
4.4. Analytical RP-HPLC of Peptides
4.5. Circular Dichroism Spectroscopy
4.6. Measurement of Anticancer Activity
4.7. Measurement of Hemolytic Activity
Acknowledgements
References
- Edwards, B.K.; Brown, M.L.; Wingo, P.A.; Howe, H.L.; Ward, E.; Ries, L.A.; Schrag, D.; Jamison, P.M.; Jemal, A.; Wu, X.C.; et al. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J. Natl. Cancer Inst 2005, 97, 1407–1427. [Google Scholar]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin 2011, 61, 69–90. [Google Scholar]
- Hoskin, D.W.; Ramamoorthy, A. Studies on anticancer activities of antimicrobial peptides. Biochim. Biophys. Acta 2008, 1778, 357–375. [Google Scholar]
- Eisenhofer, G.; Siegert, G.; Kotzerke, J.; Bornstein, S.R.; Pacak, K. Current progress and future challenges in the biochemical diagnosis and treatment of pheochromocytomas and paragangliomas. Horm. Metab. Res 2008, 40, 329–337. [Google Scholar]
- Dennison, S.R.; Whittaker, M.; Harris, F.; Phoenix, D.A. Anticancer alpha-helical peptides and structure/function relationships underpinning their interactions with tumour cell membranes. Curr. Protein Pept. Sci 2006, 7, 487–499. [Google Scholar]
- Papo, N.; Shai, Y. Host defense peptides as new weapons in cancer treatment. Cell Mol. Life Sci 2005, 62, 784–790. [Google Scholar]
- Ran, S.; Downes, A.; Thorpe, P.E. Increased exposure of anionic phospholipids on the surface of tumor blood vessels. Cancer Res 2002, 62, 6132–6140. [Google Scholar]
- Yoon, W.H.; Park, H.D.; Lim, K.; Hwang, B.D. Effect of O-glycosylated mucin on invasion and metastasis of HM7 human colon cancer cells. Biochem. Biophys. Res. Commun 1996, 222, 694–699. [Google Scholar]
- Sok, M.; Sentjurc, M.; Schara, M. Membrane fluidity characteristics of human lung cancer. Cancer Lett 1999, 139, 215–220. [Google Scholar]
- Chaudhary, J.; Munshi, M. Scanning electron microscopic analysis of breast aspirates. Cytopathology 1995, 6, 162–167. [Google Scholar]
- Huang, Y.B.; Wang, X.F.; Wang, H.Y.; Liu, Y.; Chen, Y. Studies on mechanism of action of anticancer peptides by modulation of hydrophobicity within a defined structural framework. Mol. Cancer Ther 2011, 10, 416–426. [Google Scholar]
- Chen, Y.; Mant, C.T.; Farmer, S.W.; Hancock, R.E.; Vasil, M.L.; Hodges, R.S. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem 2005, 280, 12316–12329. [Google Scholar]
- Chen, Y.; Mant, C.T.; Hodges, R.S. Determination of stereochemistry stability coefficients of amino acid side-chains in an amphipathic alpha-helix. J. Pept. Res 2002, 59, 18–33. [Google Scholar]
- Zhou, N.E.; Mant, C.T.; Hodges, R.S. Effect of preferred binding domains on peptide retention behavior in reversed-phase chromatography: Amphipathic alpha-helices. Pept. Res 1990, 3, 8–20. [Google Scholar]
- Kovacs, J.M.; Mant, C.T.; Hodges, R.S. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or conformational effects. Biopolymers 2006, 84, 283–297. [Google Scholar]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother 2007, 51, 1398–1406. [Google Scholar]
- Oren, Z.; Hong, J.; Shai, Y. A repertoire of novel antibacterial diastereomeric peptides with selective cytolytic activity. J. Biol. Chem 1997, 272, 14643–14649. [Google Scholar]
- Papo, N.; Shai, Y. New lytic peptides based on the d,l-amphipathic helix motif preferentially kill tumor cells compared to normal cells. Biochemistry 2003, 42, 9346–9354. [Google Scholar]
- Papo, N.; Shahar, M.; Eisenbach, L.; Shai, Y. A novel lytic peptide composed of dl-amino acids selectively kills cancer cells in culture and in mice. J. Biol. Chem 2003, 278, 21018–21023. [Google Scholar]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar]
- Chen, Y.; Vasil, A.I.; Rehaume, L.; Mant, C.T.; Burns, J.L.; Vasil, M.L.; Hancock, R.E.; Hodges, R.S. Comparison of biophysical and biologic properties of alpha-helical enantiomeric antimicrobial peptides. Chem. Biol. Drug Des 2006, 67, 162–173. [Google Scholar]
- Huang, J.F.; Xu, Y.M.; Hao, D.M.; Huang, Y.B.; Liu, Y.; Chen, Y.X. Structure-guided de novo design of α-helical antimicrobial peptide with enhanced specificity. Pure Appl. Chem 2010, 82, 243–257. [Google Scholar]

| Group | No. | Peptide | Amino Acid Sequencea |
|---|---|---|---|
| Parent | 1 | P | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| Polar face group | 2 | K7D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| 3 | K14D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 4 | K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 5 | K7D/K14D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 6 | K14D/K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 7 | K7D/K14D/K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 8 | K7D/K10D/K14D/K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 9 | K3D/K7D/K10D/K14D/K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 10 | K1D/K3D/K7D/K10D/K14D/K22D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| Non-polar face group | 11 | L6D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide |
| 12 | L12D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 13 | L20D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 14 | L6D/L12D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 15 | L12D/L20D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 16 | L6D/L12D/L20D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 17 | L6D/L12D/L17D/L20D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| 18 | L6D/L12D/L17D/L20D/L21D | Ac-K-W-K-S-F-L-K-T-F-K-S-L-K-K-T-V-L-H-T-L-L-K-A-I-S-S-amide | |
| Peptides a | tR (min) b 25 °C | Benign c | 50% TFE d | ||
|---|---|---|---|---|---|
| [θ]222 | % helix e | [θ]222 | % helix e | ||
| P | 46.9 | −14550 | 36.66 | −39700 | 100.00 |
| K7D | 44.1 | −6050 | 15.22 | −30900 | 77.77 |
| K14D | 43.5 | −15550 | 39.16 | −35250 | 88.72 |
| K22D | 43.3 | −8400 | 21.14 | −33950 | 85.48 |
| K7D/K14 D | 40.9 | −8750 | 22.02 | −28450 | 71.65 |
| K14D/K22D | 40.1 | −6350 | 15.95 | −32150 | 81.00 |
| K7D/K14D/K22D | 37.9 | −5000 | 12.59 | −26350 | 66.33 |
| K7D/K10D/K14D/K22D | 35.5 | −7350 | 22.48 | −19400 | 48.84 |
| K3D/K7D/K10D/K14D/K22D | 34.6 | −4750 | 14.59 | −21400 | 53.99 |
| K1D/K3D/K7D/K10D/K14D/K22D | 34.6 | −5800 | 17.74 | −19300 | 48.61 |
| L6D | 44.6 | −8500 | 21.45 | −39550 | 99.61 |
| L12D | 42.9 | −8350 | 21.01 | −37750 | 95.08 |
| L20D | 42.5 | −6900 | 17.42 | −36900 | 92.90 |
| L6D/L12D | 40.8 | −6800 | 17.14 | −29050 | 73.13 |
| L12D/L20D | 37.8 | −5550 | 13.93 | −30050 | 75.69 |
| L6D/L12D/L20D | 37.4 | −4950 | 12.45 | −28450 | 71.66 |
| L6D/L12D/L17D/L20D | 36.3 | −4050 | 12.46 | −15050 | 37.88 |
| L6D/L12D/L17D/L20D/L21D | 34.8 | −3600 | 10.97 | −13150 | 33.20 |
| Peptides a | MHC b (μmol/L) | IC50 c (μmol/L) | Therapeutic Index d | Fold e |
|---|---|---|---|---|
| P | 5.20 ± 0.02 | 1.71 ± 0.07 | 3.04 | 1.0 |
| K7D | 10.4 ± 0.04 | 1.29 ± 0.03 | 8.07 | 2.7 |
| K14D | 5.20 ± 0.02 | 1.52 ± 0.05 | 3.42 | 1.1 |
| K22D | 20.81 ± 0.10 | 1.39 ± 0.02 | 14.97 | 4.9 |
| K7D/K14D | 20.81 ± 0.15 | 2.06 ± 0.01 | 10.10 | 3.3 |
| K14D/K22D | 20.81 ± 0.06 | 1.40 ± 0.09 | 14.86 | 4.9 |
| K7D/K14D/K22D | 81.31 ± 0.17 | 4.45 ± 0.19 | 18.27 | 6.0 |
| K7D/K10D/K14D/K22D | 325.20 ± 0.82 | 4.79 ± 0.23 | 67.89 | 22.3 |
| K3D/K7D/K10D/K14D/K22D | >325.20 | 20.41 ± 0.64 | 31.87 | 10.5 |
| K1D/K3D/K7D/K10D/K14D/K22D | >325.20 | 18.07 ± 0.48 | 35.99 | 11.8 |
| L6D | 10.40 ± 0.08 | 2.23 ± 0.10 | 4.67 | 1.5 |
| L12D | 20.81 ± 0.03 | 2.63 ± 0.07 | 7.91 | 2.6 |
| L20D | 20.81 ± 0.13 | 2.33 ± 0.06 | 8.93 | 2.9 |
| L6D/L12D | 20.81 ± 0.10 | 3.01 ± 0.08 | 6.91 | 2.3 |
| L12D/L20D | 81.31 ± .43 | 3.14 ± 0.06 | 25.89 | 8.5 |
| L6D/L12D/L20D | 162.61 ± 0.19 | 7.80 ± 0.26 | 20.85 | 6.9 |
| L6D/L12D/L17D/L20D | 81.31 ± 1.05 | 9.69 ± 0.38 | 8.39 | 2.8 |
| L6D/L12D/L17D/L20D/L21D | >325.20 | 12.88 ± 0.15 | 50.50 | 16.6 |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Huang, Y.-B.; He, L.-Y.; Jiang, H.-Y.; Chen, Y.-X. Role of Helicity on the Anticancer Mechanism of Action of Cationic-Helical Peptides. Int. J. Mol. Sci. 2012, 13, 6849-6862. https://doi.org/10.3390/ijms13066849
Huang Y-B, He L-Y, Jiang H-Y, Chen Y-X. Role of Helicity on the Anticancer Mechanism of Action of Cationic-Helical Peptides. International Journal of Molecular Sciences. 2012; 13(6):6849-6862. https://doi.org/10.3390/ijms13066849
Chicago/Turabian StyleHuang, Yi-Bing, Li-Yan He, Hong-Yu Jiang, and Yu-Xin Chen. 2012. "Role of Helicity on the Anticancer Mechanism of Action of Cationic-Helical Peptides" International Journal of Molecular Sciences 13, no. 6: 6849-6862. https://doi.org/10.3390/ijms13066849





