Biogenesis and Mechanism of Action of Small Non-Coding RNAs: Insights from the Point of View of Structural Biology
Abstract
:1. Introduction
2. Micro-RNAs (miRNAs)
3. piRNAs
4. siRNAs
5. Small Non-Coding RNA Processors
5.1. Drosha and the Nuclear Microprocessor
5.2. Cytoplasmic Processors: Dicer
6. Efectors of the Small Non-Coding RNA Regulatory Networks: Proteins from the Argonaute Family
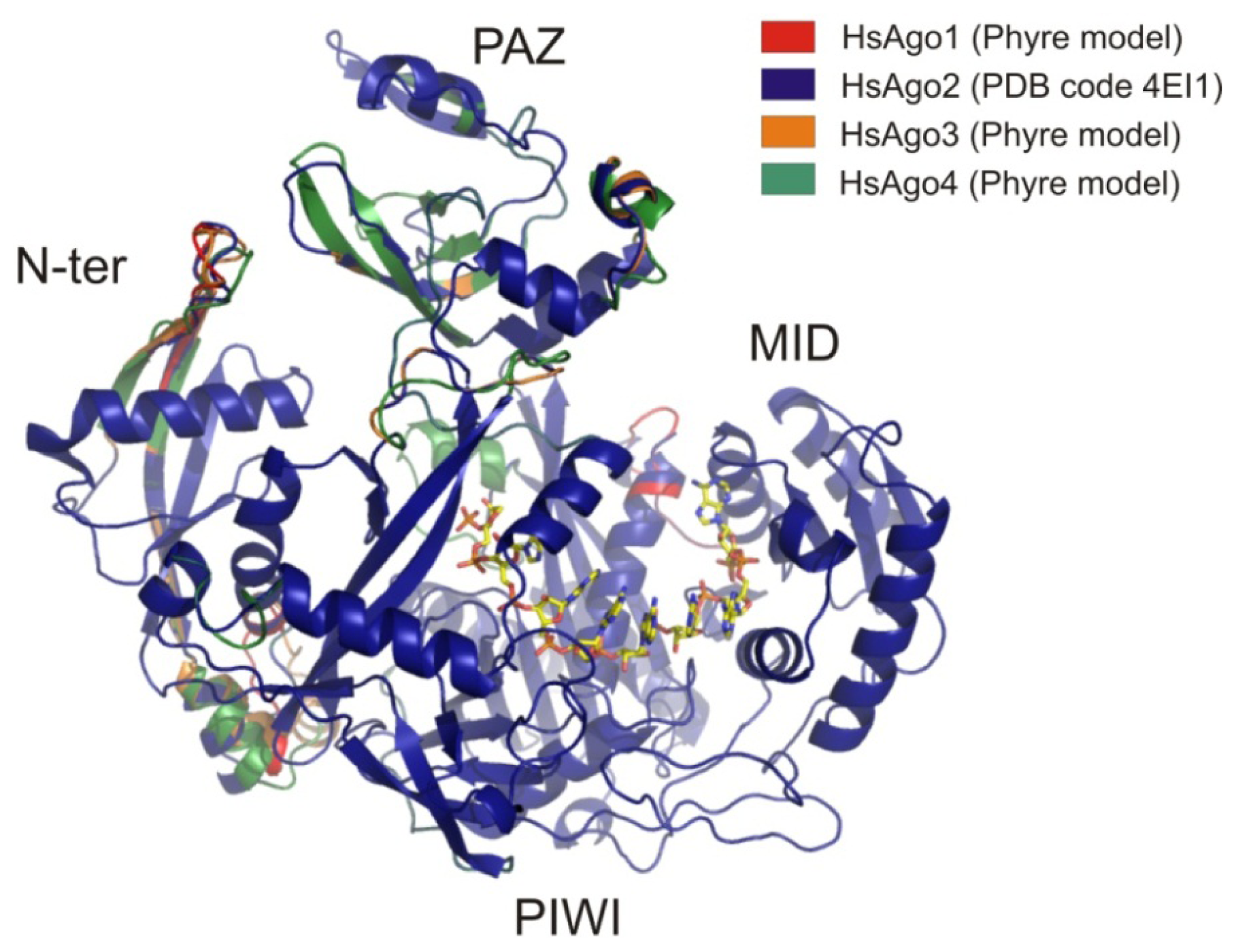
6.1. AGO Sub-Family
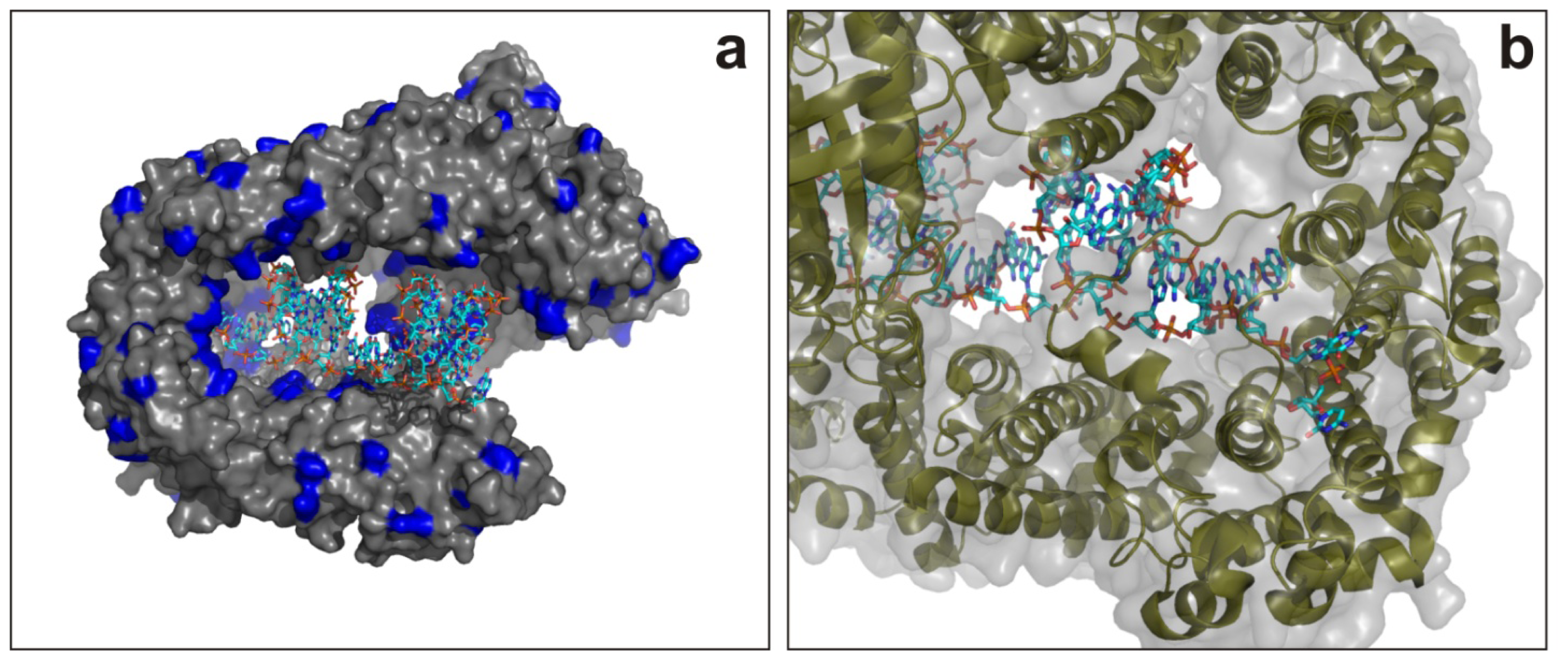
6.1.1. AGO2
6.1.2. Other Members of the AGO Sub-Family
7. Helper Proteins and Additional Members of the Non-Coding RNA Effector Complexes
7.1. TRBP
7.2. GW182
7.3. PIWI Proteins
8. Other Helper Proteins: Exportin 5
9. Conclusions
Acknowledgments
References
- Maniatis, T.; Tasic, B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 2002, 418, 236–243. [Google Scholar]
- Hui, J.; Bindereif, A. Alternative pre-mRNA splicing in the human system: Unexpected role of repetitive sequences as regulatory elements. Biol. Chem 2005, 386, 1265–1271. [Google Scholar]
- Smith, C.W.; Valcarcel, J. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci 2000, 25, 381–388. [Google Scholar]
- Boue, S.; Letunic, I.; Bork, P. Alternative splicing and evolution. Bioessays 2003, 25, 1031–1034. [Google Scholar]
- Mehler, M.F.; Mattick, J.S. Non-coding RNAs in the nervous system. J. Physiol 2006, 575, 333–341. [Google Scholar]
- Louro, R.; Nakaya, H.I.; Amaral, P.P.; Festa, F.; Sogayar, M.C.; da Silva, A.M.; Verjovski-Almeida, S.; Reis, E.M. Androgen responsive intronic non-coding RNAs. BMC Biol 2007, 5. [Google Scholar] [CrossRef] [Green Version]
- Tomaru, Y.; Hayashizaki, Y. Cancer research with non-coding RNA. Cancer Sci 2006, 97, 1285–1290. [Google Scholar]
- Chen, Z.; Zhang, J.; Kong, J.; Li, S.; Fu, Y.; Li, S.; Zhang, H.; Li, Y.; Zhu, Y. Diversity of endogenous small non-coding RNAs in Oryza sativa. Genetica 2006, 128, 21–31. [Google Scholar]
- Mattick, J.S.; Makunin, I.V. Non-coding RNA. Hum. Mol. Genet 2006, 15, R17–R29. [Google Scholar]
- Missal, K.; Rose, D.; Stadler, P.F. Non-coding RNAs in Ciona intestinalis. Bioinformatics 2005, 21 Suppl 2, ii77–ii78. [Google Scholar]
- Royo, H.; Bortolin, M.L.; Seitz, H.; Cavaille, J. Small non-coding RNAs and genomic imprinting. Cytogenet. Genome Res 2006, 113, 99–108. [Google Scholar]
- Eckstein, F. Small non-coding RNAs as magic bullets. Trends Biochem. Sci 2005, 30, 445–452. [Google Scholar]
- Murchison, E.P.; Hannon, G.J. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr. Opin. Cell Biol 2004, 16, 223–229. [Google Scholar]
- Faehnle, C.R.; Joshua-Tor, L. Argonautes confront new small RNAs. Curr. Opin. Chem. Biol 2007, 11, 569–577. [Google Scholar]
- Perron, M.P.; Provost, P. Protein components of the microRNA pathway and human diseases. Methods Mol. Biol 2009, 487, 369–385. [Google Scholar]
- Patel, D.J.; Ma, J.B.; Yuan, Y.R.; Ye, K.; Pei, Y.; Kuryavyi, V.; Malinina, L.; Meister, G.; Tuschl, T. Structural biology of RNA silencing and its functional implications. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 81–93. [Google Scholar]
- Ji, X. The mechanism of RNase III action: How dicer dices. Curr. Top. Microbiol. Immunol 2008, 320, 99–116. [Google Scholar]
- Wan, Y.; Kertesz, M.; Spitale, R.C.; Segal, E.; Chang, H.Y. Understanding the transcriptome through RNA structure. Nat. Rev. Genet 2011, 12, 641–655. [Google Scholar]
- Davis-Dusenbery, B.N.; Hata, A. Mechanisms of control of microRNA biogenesis. J. Biochem 2010, 148, 381–392. [Google Scholar]
- Newman, M.A.; Hammond, S.M. Emerging paradigms of regulated microRNA processing. Genes Dev 2010, 24, 1086–1092. [Google Scholar]
- Lee, Y.; Han, J.; Yeom, K.H.; Jin, H.; Kim, V.N. Drosha in primary microRNA processing. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 51–57. [Google Scholar]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 2004, 18, 3016–3027. [Google Scholar]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Di Leva, G.; Calin, G.A.; Croce, C.M. MicroRNAs: Fundamental facts and involvement in human diseases. Birth Defects Res. C Embryo. Today 2006, 78, 180–189. [Google Scholar]
- De, N.; Macrae, I.J. Purification and assembly of human Argonaute, Dicer, and TRBP complexes. Methods Mol. Biol 2011, 725, 107–119. [Google Scholar]
- Ye, X.; Huang, N.; Liu, Y.; Paroo, Z.; Huerta, C.; Li, P.; Chen, S.; Liu, Q.; Zhang, H. Structure of C3PO and mechanism of human RISC activation. Nat. Struct. Mol. Biol 2011, 18, 650–657. [Google Scholar]
- Krutzfeldt, J.; Stoffel, M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab 2006, 4, 9–12. [Google Scholar]
- Maroney, P.A.; Yu, Y.; Nilsen, T.W. MicroRNAs, mRNAs, and translation. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 531–535. [Google Scholar]
- Osada, H.; Takahashi, T. MicroRNAs in biological processes and carcinogenesis. Carcinogenesis 2007, 28, 2–12. [Google Scholar]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol 2006, 57, 19–53. [Google Scholar]
- Zhang, B.; Wang, Q.; Pan, X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol 2007, 210, 279–289. [Google Scholar]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J 2007, 26, 775–783. [Google Scholar]
- Lin, S.L.; Miller, J.D.; Ying, S.Y. Intronic microRNA (miRNA). J. Biomed. Biotechnol 2006, 2006. [Google Scholar] [CrossRef]
- De Yebenes, V.G.; Belver, L.; Pisano, D.G.; Gonzalez, S.; Villasante, A.; Croce, C.; He, L.; Ramiro, A.R. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med 2008, 205, 2199–2206. [Google Scholar]
- Di Leva, G.; Gasparini, P.; Piovan, C.; Ngankeu, A.; Garofalo, M.; Taccioli, C.; Iorio, M.V.; Li, M.; Volinia, S.; Alder, H.; et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J. Natl. Cancer Inst 2010, 102, 706–721. [Google Scholar]
- Georges, S.A.; Biery, M.C.; Kim, S.Y.; Schelter, J.M.; Guo, J.; Chang, A.N.; Jackson, A.L.; Carleton, M.O.; Linsley, P.S.; Cleary, M.A.; et al. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res 2008, 68, 10105–10112. [Google Scholar]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, 539–543. [Google Scholar]
- Grivna, S.T.; Pyhtila, B.; Lin, H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 13415–13420. [Google Scholar]
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 2006, 20, 1709–1714. [Google Scholar]
- Kirino, Y.; Mourelatos, Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 2007, 13, 1397–1401. [Google Scholar]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar]
- Lau, N.C.; Robine, N.; Martin, R.; Chung, W.J.; Niki, Y.; Berezikov, E.; Lai, E.C. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 2009, 19, 1776–1785. [Google Scholar]
- He, Z.; Kokkinaki, M.; Pant, D.; Gallicano, G.I.; Dym, M. Small RNA molecules in the regulation of spermatogenesis. Reproduction 2009, 137, 901–911. [Google Scholar]
- Couvillion, M.T.; Lee, S.R.; Hogstad, B.; Malone, C.D.; Tonkin, L.A.; Sachidanandam, R.; Hannon, G.J.; Collins, K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev 2009, 23, 2016–2032. [Google Scholar]
- Castaneda, J.; Genzor, P.; Bortvin, A. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res 2011, 714, 95–104. [Google Scholar]
- Kawaoka, S.; Izumi, N.; Katsuma, S.; Tomari, Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell 2011, 43, 1015–1022. [Google Scholar]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol 2011, 12, 246–258. [Google Scholar]
- Zhang, C. Novel functions for small RNA molecules. Curr. Opin. Mol. Ther 2009, 11, 641–651. [Google Scholar]
- Nishida, K.M.; Okada, T.N.; Kawamura, T.; Mituyama, T.; Kawamura, Y.; Inagaki, S.; Huang, H.; Chen, D.; Kodama, T.; Siomi, H.; Siomi, M.C. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 2009, 28, 3820–3831. [Google Scholar]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol 2009, 10, 126–139. [Google Scholar]
- Malone, C.D.; Hannon, G.J. Small RNAs as guardians of the genome. Cell 2009, 136, 656–668. [Google Scholar]
- Grimson, A.; Srivastava, M.; Fahey, B.; Woodcroft, B.J.; Chiang, H.R.; King, N.; Degnan, B.M.; Rokhsar, D.S.; Bartel, D.P. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 2008, 455, 1193–1197. [Google Scholar]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar]
- Sijen, T.; Steiner, F.A.; Thijssen, K.L.; Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 2007, 315, 244–247. [Google Scholar]
- Samaha, H.; Delorme, V.; Pontvianne, F.; Cooke, R.; Delalande, F.; van Dorsselaer, A.; Echeverria, M.; Saez-Vasquez, J. Identification of protein factors and U3 snoRNAs from a Brassica oleracea RNP complex involved in the processing of pre-rRNA. Plant J 2010, 61, 383–398. [Google Scholar]
- Maniar, J.M.; Fire, A.Z. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr. Biol 2011, 21, 449–459. [Google Scholar]
- Tam, O.H.; Aravin, A.A.; Stein, P.; Girard, A.; Murchison, E.P.; Cheloufi, S.; Hodges, E.; Anger, M.; Sachidanandam, R.; Schultz, R.M.; Hannon, G.J. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008, 453, 534–538. [Google Scholar]
- Correa, R.L.; Steiner, F.A.; Berezikov, E.; Ketting, R.F. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet 2010, 6. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Liu, J.; Kiba, T.; Woo, J.; Ojo, T.; Hafner, M.; Tuschl, T.; Chua, N.H.; Wang, X.J. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 2011, 67, 292–304. [Google Scholar]
- Chellappan, P.; Xia, J.; Zhou, X.; Gao, S.; Zhang, X.; Coutino, G.; Vazquez, F.; Zhang, W.; Jin, H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 2010, 38, 6883–6894. [Google Scholar]
- Chen, X. Small RNAs in development—Insights from plants. Curr. Opin. Genet. Dev 2012, in press. [Google Scholar]
- Soifer, H.S.; Zaragoza, A.; Peyvan, M.; Behlke, M.A.; Rossi, J.J. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res 2005, 33, 846–856. [Google Scholar]
- Aporntewan, C.; Phokaew, C.; Piriyapongsa, J.; Ngamphiw, C.; Ittiwut, C.; Tongsima, S.; Mutirangura, A. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One 2011, 6. [Google Scholar] [CrossRef]
- Qi, H.; Watanabe, T.; Ku, H.Y.; Liu, N.; Zhong, M.; Lin, H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem 2011, 286, 3789–3797. [Google Scholar]
- Costa, F.F. Non-coding RNAs, epigenetics and complexity. Gene 2008, 410, 9–17. [Google Scholar]
- Nowotny, M.; Yang, W. Structural and functional modules in RNA interference. Curr. Opin. Struct. Biol 2009, 19, 286–293. [Google Scholar]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol 2012, 19, 436–440. [Google Scholar]
- Perron, M.P.; Provost, P. Protein interactions and complexes in human microRNA biogenesis and function. Front Biosci 2008, 13, 2537–2547. [Google Scholar]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar]
- Gregory, R.I.; Chendrimada, T.P.; Shiekhattar, R. MicroRNA biogenesis: Isolation and characterization of the microprocessor complex. Methods Mol. Biol 2006, 342, 33–47. [Google Scholar]
- Triboulet, R.; Gregory, R.I. Autoregulatory mechanisms controlling the microprocessor. Adv. Exp. Med. Biol 2010, 700, 56–66. [Google Scholar]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol 2003, 13, 784–789. [Google Scholar]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; Kim, V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar]
- de la Chapelle, A.; Herva, R.; Koivisto, M.; Aula, P. A deletion in chromosome 22 can cause DiGeorge syndrome. Hum. Genet 1981, 57, 253–256. [Google Scholar]
- Kelley, R.I.; Zackai, E.H.; Emanuel, B.S.; Kistenmacher, M.; Greenberg, F.; Punnett, H.H. The association of the DiGeorge anomalad with partial monosomy of chromosome 22. J. Pediatr 1982, 101, 197–200. [Google Scholar]
- Mueller, G.A.; Miller, M.T.; Derose, E.F.; Ghosh, M.; London, R.E.; Hall, T.M. Solution structure of the Drosha double-stranded RNA-binding domain. Silence 2010, 1. [Google Scholar] [CrossRef]
- Senturia, R.; Faller, M.; Yin, S.; Loo, J.A.; Cascio, D.; Sawaya, M.R.; Hwang, D.; Clubb, R.T.; Guo, F. Structure of the dimerization domain of DiGeorge critical region 8. Protein Sci 2010, 19, 1354–1365. [Google Scholar]
- Sohn, S.Y.; Bae, W.J.; Kim, J.J.; Yeom, K.H.; Kim, V.N.; Cho, Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol 2007, 14, 847–853. [Google Scholar]
- Faller, M.; Matsunaga, M.; Yin, S.; Loo, J.A.; Guo, F. Heme is involved in microRNA processing. Nat. Struct. Mol. Biol 2007, 14, 23–29. [Google Scholar]
- Barr, I.; Smith, A.T.; Senturia, R.; Chen, Y.; Scheidemantle, B.D.; Burstyn, J.N.; Guo, F. DiGeorge critical region 8 (DGCR8) is a double-cysteine-ligated heme protein. J. Biol. Chem 2011, 286, 16716–16725. [Google Scholar]
- Barr, I.; Smith, A.T.; Chen, Y.; Senturia, R.; Burstyn, J.N.; Guo, F. Ferric, not ferrous, heme activates RNA-binding protein DGCR8 for primary microRNA processing. Proc. Natl. Acad. Sci. USA 2012, 109, 1919–1924. [Google Scholar]
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 2002, 21, 5875–5885. [Google Scholar]
- Pellino, J.L.; Jaskiewicz, L.; Filipowicz, W.; Sontheimer, E.J. ATP modulates siRNA interactions with an endogenous human Dicer complex. RNA 2005, 11, 1719–1724. [Google Scholar]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett 2005, 579, 5822–5829. [Google Scholar]
- Macrae, I.J.; Li, F.; Zhou, K.; Cande, W.Z.; Doudna, J.A. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 73–80. [Google Scholar]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol 2007, 14, 934–940. [Google Scholar]
- Cook, A.; Conti, E. Dicer measures up. Nat. Struct. Mol. Biol 2006, 13, 190–192. [Google Scholar]
- Welker, N.C.; Maity, T.S.; Ye, X.; Aruscavage, P.J.; Krauchuk, A.A.; Liu, Q.; Bass, B.L. Dicer’s helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol. Cell 2011, 41, 589–599. [Google Scholar]
- Park, J.E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar]
- Du, Z.; Lee, J.K.; Tjhen, R.; Stroud, R.M.; James, T.L. Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage. Proc. Natl. Acad. Sci. USA 2008, 105, 2391–2396. [Google Scholar]
- Makarova, K.S.; Wolf, Y.I.; van der Oost, J.; Koonin, E.V. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 2009, 4. [Google Scholar] [CrossRef]
- Ekwall, K. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell 2004, 13, 304–305. [Google Scholar]
- Verdel, A.; Vavasseur, A.; Le Gorrec, M.; Touat-Todeschini, L. Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol 2009, 53, 245–257. [Google Scholar]
- Miyoshi, K.; Tsukumo, H.; Nagami, T.; Siomi, H.; Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 2005, 19, 2837–2848. [Google Scholar]
- Verdel, A.; Jia, S.; Gerber, S.; Sugiyama, T.; Gygi, S.; Grewal, S.I.; Moazed, D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004, 303, 672–676. [Google Scholar]
- Mescalchin, A.; Detzer, A.; Weirauch, U.; Hahnel, M.J.; Engel, C.; Sczakiel, G. Antisense tools for functional studies of human Argonaute proteins. RNA 2010, 16, 2529–2536. [Google Scholar]
- Azuma-Mukai, A.; Oguri, H.; Mituyama, T.; Qian, Z.R.; Asai, K.; Siomi, H.; Siomi, M.C. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. USA 2008, 105, 7964–7969. [Google Scholar]
- Wang, D.; Zhang, Z.; O’Loughlin, E.; Lee, T.; Houel, S.; O’Carroll, D.; Tarakhovsky, A.; Ahn, N.G.; Yi, R. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 2012, 26, 693–704. [Google Scholar]
- Song, J.J.; Smith, S.K.; Hannon, G.J.; Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 2004, 305, 1434–1437. [Google Scholar]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Tuschl, T.; Patel, D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 2008, 456, 921–926. [Google Scholar]
- Wang, Y.; Sheng, G.; Juranek, S.; Tuschl, T.; Patel, D.J. Structure of the guide-strand-containing argonaute silencing complex. Nature 2008, 456, 209–213. [Google Scholar]
- Yuan, Y.R.; Pei, Y.; Ma, J.B.; Kuryavyi, V.; Zhadina, M.; Meister, G.; Chen, H.Y.; Dauter, Z.; Tuschl, T.; Patel, D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 2005, 19, 405–419. [Google Scholar]
- Song, J.J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol 2003, 10, 1026–1032. [Google Scholar]
- Schirle, N.T.; Macrae, I.J. The crystal structure of human Argonaute2. Science 2012, 336, 1037–1040. [Google Scholar]
- Boland, A.; Huntzinger, E.; Schmidt, S.; Izaurralde, E.; Weichenrieder, O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc. Natl. Acad. Sci. USA 2011, 108, 10466–10471. [Google Scholar]
- Wang, Y.; Juranek, S.; Li, H.; Sheng, G.; Wardle, G.S.; Tuschl, T.; Patel, D.J. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 2009, 461, 754–761. [Google Scholar]
- Lima, W.F.; Wu, H.; Nichols, J.G.; Sun, H.; Murray, H.M.; Crooke, S.T. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem 2009, 284, 26017–26028. [Google Scholar]
- Zeng, Y.; Sankala, H.; Zhang, X.; Graves, P.R. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J 2008, 413, 429–436. [Google Scholar]
- Ma, J.B.; Ye, K.; Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004, 429, 318–322. [Google Scholar]
- Maiti, R.; van Domselaar, G.H.; Zhang, H.; Wishart, D.S. SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res 2004, 32, W590–W594. [Google Scholar]
- Wang, H.W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol 2009, 16, 1148–1153. [Google Scholar]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar]
- Takimoto, K.; Wakiyama, M.; Yokoyama, S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA 2009, 15, 1078–1089. [Google Scholar]
- Wang, B.; Li, S.; Qi, H.H.; Chowdhury, D.; Shi, Y.; Novina, C.D. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat. Struct. Mol. Biol 2009, 16, 1259–1266. [Google Scholar]
- Chu, Y.; Yue, X.; Younger, S.T.; Janowski, B.A.; Corey, D.R. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res 2010, 38, 7736–7748. [Google Scholar]
- Zilberman, D.; Cao, X.; Jacobsen, S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 2003, 299, 716–719. [Google Scholar]
- Zilberman, D.; Cao, X.; Johansen, L.K.; Xie, Z.; Carrington, J.C.; Jacobsen, S.E. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol 2004, 14, 1214–1220. [Google Scholar]
- Rowley, M.J.; Avrutsky, M.I.; Sifuentes, C.J.; Pereira, L.; Wierzbicki, A.T. Independent chromatin binding of ARGONAUTE4 and SPT5L/KTF1 mediates transcriptional gene silencing. PLoS Genet 2011, 7. [Google Scholar] [CrossRef]
- Tran, R.K.; Zilberman, D.; de Bustos, C.; Ditt, R.F.; Henikoff, J.G.; Lindroth, A.M.; Delrow, J.; Boyle, T.; Kwong, S.; Bryson, T.D.; Jacobsen, S.E.; Henikoff, S. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol 2005, 6. [Google Scholar] [CrossRef]
- Broderick, J.A.; Salomon, W.E.; Ryder, S.P.; Aronin, N.; Zamore, P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA 2011, 17, 1858–1869. [Google Scholar]
- Bennett-Lovsey, R.M.; Herbert, A.D.; Sternberg, M.J.; Kelley, L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 2008, 70, 611–625. [Google Scholar]
- Kelley, L.A.; Sternberg, M.J. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc 2009, 4, 363–371. [Google Scholar]
- Parker, J.S.; Roe, S.M.; Barford, D. Molecular mechanism of target RNA transcript recognition by Argonaute-guide complexes. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 45–50. [Google Scholar]
- Reddy, T.R.; Suhasini, M.; Rappaport, J.; Looney, D.J.; Kraus, G.; Wong-Staal, F. Molecular cloning and characterization of a TAR-binding nuclear factor from T cells. AIDS Res. Hum. Retrovir 1995, 11, 663–669. [Google Scholar]
- Gatignol, A.; Buckler-White, A.; Berkhout, B.; Jeang, K.T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 1991, 251, 1597–1600. [Google Scholar]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA 2008, 105, 512–517. [Google Scholar]
- Gredell, J.A.; Dittmer, M.J.; Wu, M.; Chan, C.; Walton, S.P. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry 2010, 49, 3148–3155. [Google Scholar]
- Kalidas, S.; Sanders, C.; Ye, X.; Strauss, T.; Kuhn, M.; Liu, Q.; Smith, D.P. Drosophila R2D2 mediates follicle formation in somatic tissues through interactions with Dicer-1. Mech. Dev 2008, 125, 475–485. [Google Scholar]
- Murphy, D.; Dancis, B.; Brown, J.R. The evolution of core proteins involved in microRNA biogenesis. BMC Evol. Biol 2008, 8. [Google Scholar] [CrossRef]
- Yang, S.W.; Chen, H.Y.; Yang, J.; Machida, S.; Chua, N.H.; Yuan, Y.A. Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 2010, 18, 594–605. [Google Scholar]
- Lau, P.W.; Potter, C.S.; Carragher, B.; MacRae, I.J. Structure of the human Dicer-TRBP complex by electron microscopy. Structure 2009, 17, 1326–1332. [Google Scholar]
- Chekulaeva, M.; Parker, R.; Filipowicz, W. The GW/WG repeats of Drosophila GW182 function as effector motifs for miRNA-mediated repression. Nucleic Acids Res 2010, 38, 6673–6683. [Google Scholar]
- Lazzaretti, D.; Tournier, I.; Izaurralde, E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA 2009, 15, 1059–1066. [Google Scholar]
- Zipprich, J.T.; Bhattacharyya, S.; Mathys, H.; Filipowicz, W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA 2009, 15, 781–793. [Google Scholar]
- Fabian, M.R.; Mathonnet, G.; Sundermeier, T.; Mathys, H.; Zipprich, J.T.; Svitkin, Y.V.; Rivas, F.; Jinek, M.; Wohlschlegel, J.; Doudna, J.A.; et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell 2009, 35, 868–880. [Google Scholar]
- Jinek, M.; Fabian, M.R.; Coyle, S.M.; Sonenberg, N.; Doudna, J.A. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat. Struct. Mol. Biol 2010, 17, 238–240. [Google Scholar]
- Piao, X.; Zhang, X.; Wu, L.; Belasco, J.G. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol. Cell Biol 2010, 30, 1486–1494. [Google Scholar]
- Braun, J.E.; Huntzinger, E.; Fauser, M.; Izaurralde, E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell 2011, 44, 120–133. [Google Scholar]
- Chekulaeva, M.; Mathys, H.; Zipprich, J.T.; Attig, J.; Colic, M.; Parker, R.; Filipowicz, W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat. Struct. Mol. Biol 2011, 18, 1218–1226. [Google Scholar]
- Unhavaithaya, Y.; Hao, Y.; Beyret, E.; Yin, H.; Kuramochi-Miyagawa, S.; Nakano, T.; Lin, H. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J. Biol. Chem 2009, 284, 6507–6519. [Google Scholar]
- Siddiqi, S.; Matushansky, I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J. Cell Biochem 2012, 113, 373–380. [Google Scholar]
- Juliano, C.; Wang, J.; Lin, H. Uniting germline and stem cells: The function of PIWI proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet 2011, 45, 447–469. [Google Scholar]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar]
- Simon, B.; Kirkpatrick, J.P.; Eckhardt, S.; Reuter, M.; Rocha, E.A.; Andrade-Navarro, M.A.; Sehr, P.; Pillai, R.S.; Carlomagno, T. Recognition of 2′-O-methylated 3′-end of piRNA by the PAZ domain of a Piwi protein. Structure 2011, 19, 172–180. [Google Scholar]
- Van der Heijden, G.W.; Bortvin, A. Defending the genome in tudor style. Dev. Cell 2009, 17, 745–746. [Google Scholar]
- Liu, L.; Qi, H.; Wang, J.; Lin, H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 2011, 138, 1863–1873. [Google Scholar]
- Liu, K.; Chen, C.; Guo, Y.; Lam, R.; Bian, C.; Xu, C.; Zhao, D.Y.; Jin, J.; MacKenzie, F.; Pawson, T.; Min, J. Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain. Proc. Natl. Acad. Sci. USA 2010, 107, 18398–18403. [Google Scholar]
- Chen, C.; Jin, J.; James, D.A.; Adams-Cioaba, M.A.; Park, J.G.; Guo, Y.; Tenaglia, E.; Xu, C.; Gish, G.; Min, J.; Pawson, T. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. USA 2009, 106, 20336–20341. [Google Scholar]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003, 17, 3011–3016. [Google Scholar]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res 2004, 32, 4776–4785. [Google Scholar]
- Kim, V.N. MicroRNA precursors in motion: Exportin-5 mediates their nuclear export. Trends Cell Biol 2004, 14, 156–159. [Google Scholar]
- Lund, E.; Dahlberg, J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 59–66. [Google Scholar]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar]





© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Costa, M.C.; Leitão, A.L.; Enguita, F.J. Biogenesis and Mechanism of Action of Small Non-Coding RNAs: Insights from the Point of View of Structural Biology. Int. J. Mol. Sci. 2012, 13, 10268-10295. https://doi.org/10.3390/ijms130810268
Costa MC, Leitão AL, Enguita FJ. Biogenesis and Mechanism of Action of Small Non-Coding RNAs: Insights from the Point of View of Structural Biology. International Journal of Molecular Sciences. 2012; 13(8):10268-10295. https://doi.org/10.3390/ijms130810268
Chicago/Turabian StyleCosta, Marina C., Ana Lúcia Leitão, and Francisco J. Enguita. 2012. "Biogenesis and Mechanism of Action of Small Non-Coding RNAs: Insights from the Point of View of Structural Biology" International Journal of Molecular Sciences 13, no. 8: 10268-10295. https://doi.org/10.3390/ijms130810268





