Consequences of Morphology on Molecularly Imprinted Polymer-Ligand Recognition
Abstract
:1. Introduction
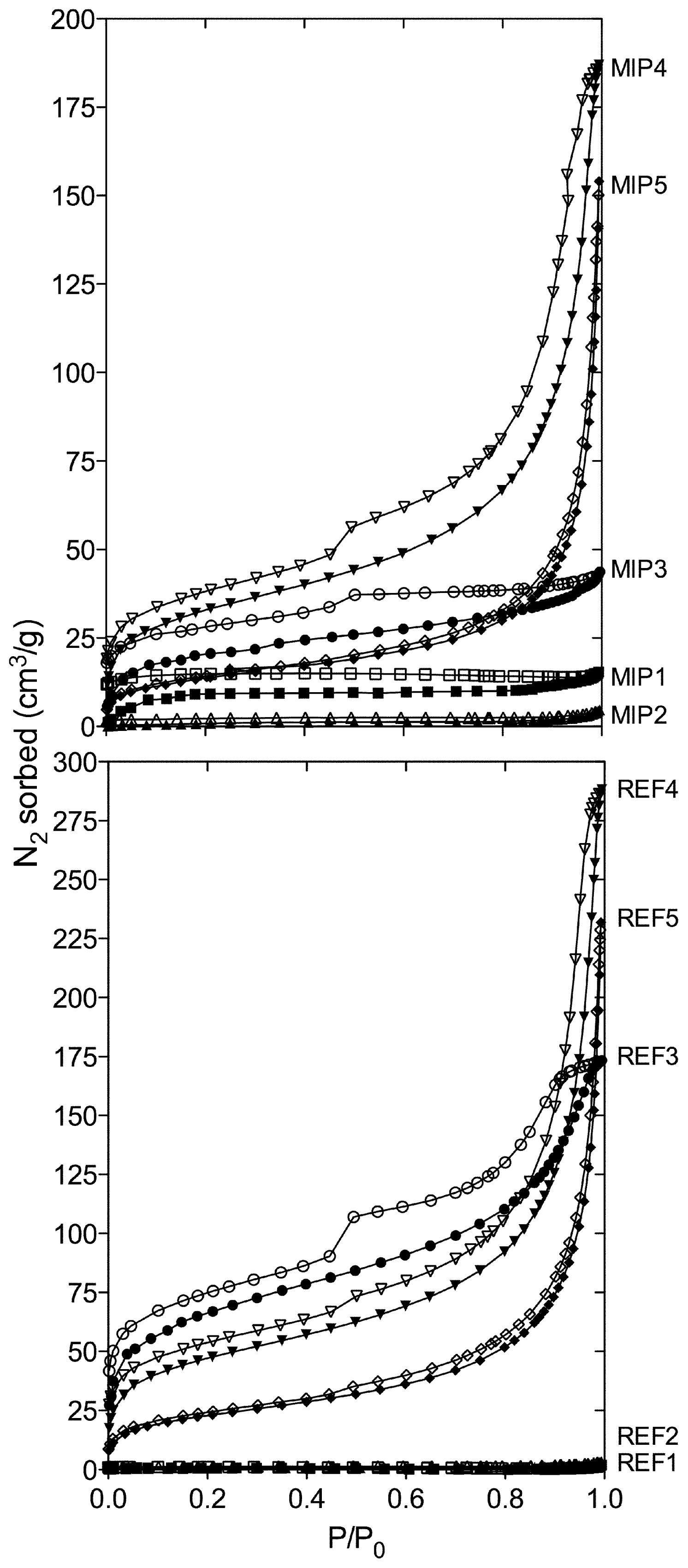
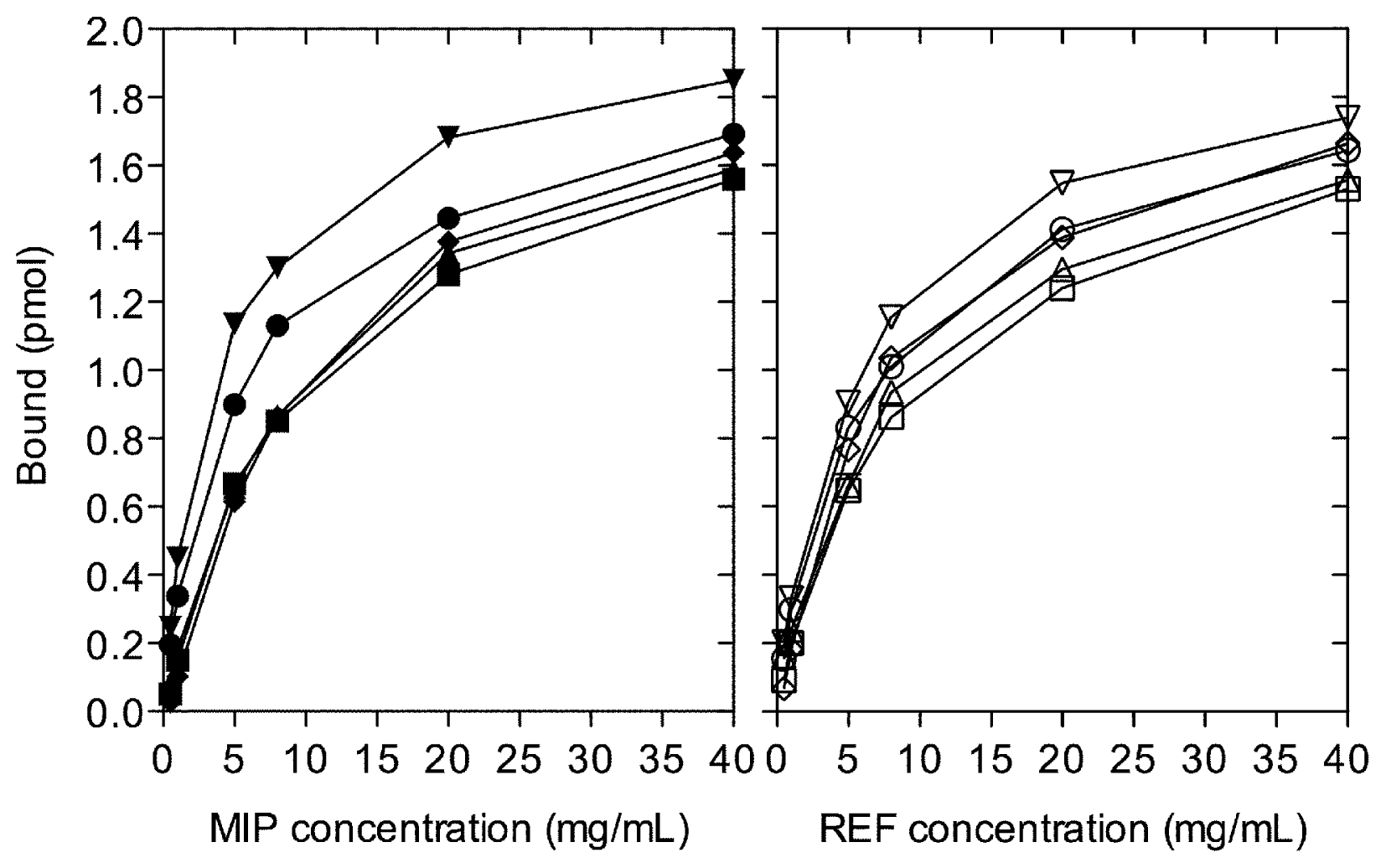
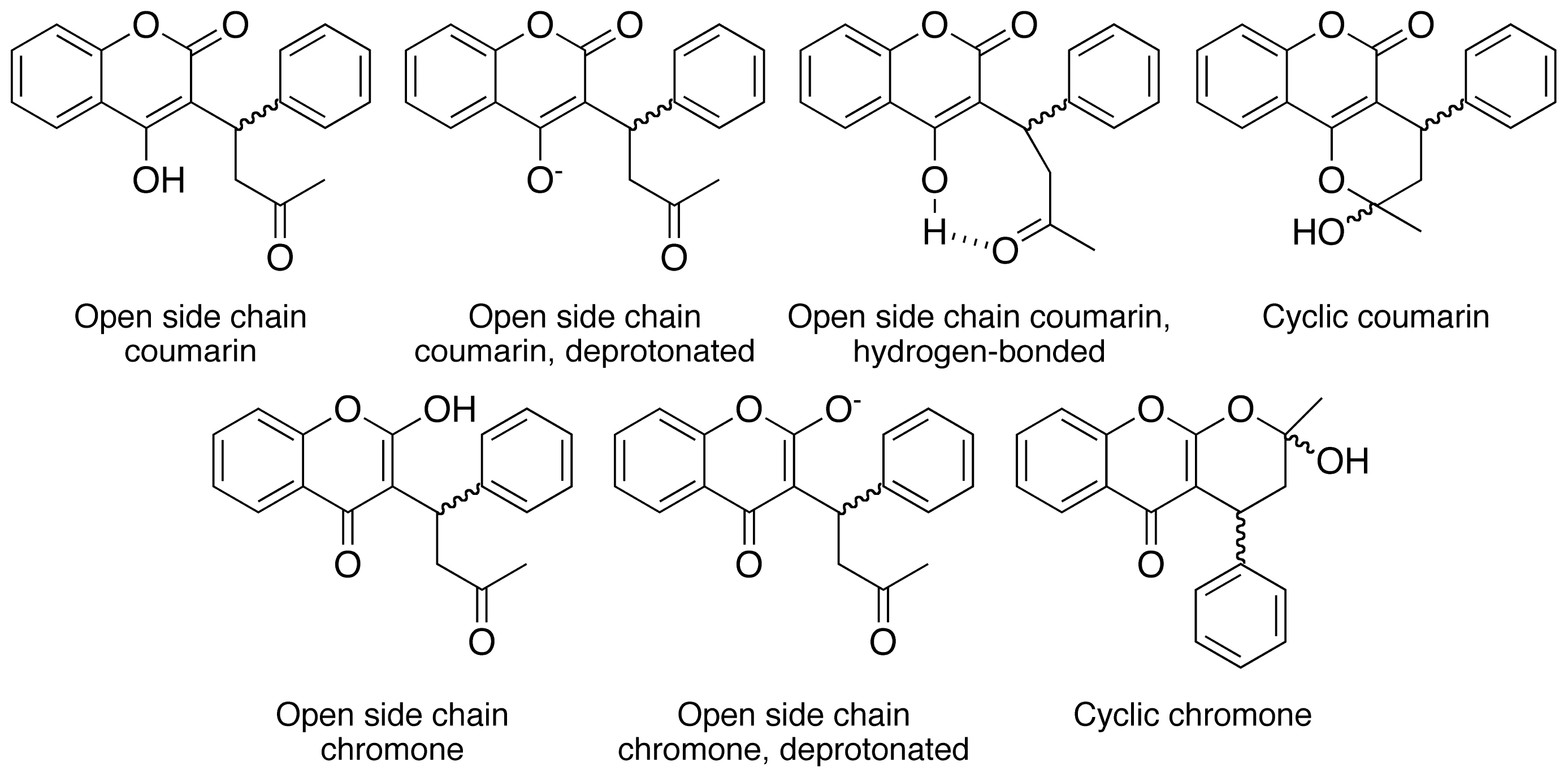
2. Results and Discussion
3. Experimental Section
3.1. Chemicals
3.2. Polymer Syntheses
3.3. Polymer Surface Area and Porosity Measurements
3.4. Radioligand Binding Studies
3.5. Swelling Studies
4. Conclusions
Acknowledgments
- Conflict of InterestThe authors declare no conflict of interest.
References
- Wulff, G. Molecular imprinting. Ann. N. Y. Acad. Sci 1984, 434, 327–333. [Google Scholar]
- Mosbach, K.; Ramström, O. The emerging technique of molecular imprinting and its future impact on biotechnology. Biotechnology 1996, 14, 163–170. [Google Scholar]
- Molecularly Imprinted Polymers: Man-Made Mimics of Antibodies and Their Applications in Analytical Chemistry; Sellergren, B. (Ed.) Elsevier: Amsterdam, The Netherlands, 2001.
- Alexander, C.; Andersson, H.S.; Andersson, L.I.; Ansell, R.J.; Kirsch, N.; Nicholls, I.A.; O’Mahony, J.; Whitcombe, M.J. Molecular imprinting science and technology: A survey of the literature for the years up to and including 2003. J. Mol. Recognit 2006, 19, 106–180. [Google Scholar]
- Sellergren, B.; Shea, K.J. Influence of polymer morphology on the ability of imprinted network polymers to resolve enantiomers. J. Chromatogr. A 1993, 635, 31–49. [Google Scholar]
- Nicholls, I. Thermodynamic considerations for the design of and ligand recognition by molecularly imprinted polymers. Chem. Lett 1995, 24, 1035–1036. [Google Scholar]
- Wu, X.; Carroll, W.R.; Shimizu, K.D. Stochastic lattice model simulations of molecularly imprinted polymers. Chem. Mater 2008, 20, 4335–4346. [Google Scholar]
- Baggiani, C.; Giovannoli, C.; Anfossi, L.; Passini, C.; Baravalle, P.; Giraudi, G. A Connection between the binding properties of imprinted and nonimprinted polymers: A change of perspective in molecular imprinting. J. Am. Chem. Soc 2012, 134, 1513–1518. [Google Scholar]
- Nicholls, I.A.; Piletsky, S.A.; Chen, B.; Chianella, I.; Turner, A.P.F. Thermodynamic Considerations and The Use of Molecular Modeling as a Tool for Predicting MIP Performance. In Molecularly Imprinted Materials; Yan, M., Ramström, O., Eds.; Marcel Dekker: New York, NY, USA, 2004. [Google Scholar]
- Karim, K.; Breton, F.; Rouillon, R.; Piletska, E.V.; Guerreiro, A.; Chianella, I.; Piletsky, S.A. How to find effective functional monomers for effective molecularly imprinted polymers? Adv. Drug Deliv. Rev 2005, 57, 1795–1808. [Google Scholar]
- Nicholls, I.A.; Andersson, H.S.; Charlton, C.; Henschel, H.; Karlsson, B.C.G.; Karlsson, J.G.; O’Mahony, J.; Rosengren, A.M.; Rosengren, K.J.; Wikman, S. Theoretical and computational strategies for rational molecularly imprinted polymer design. Biosens. Bioelectron 2009, 25, 543–552. [Google Scholar]
- Nicholls, I.; Andersson, H.; Golker, K.; Henschel, H.; Karlsson, B.; Olsson, G.; Rosengren, A.; Shoravi, S.; Suriyanarayanan, S.; Wiklander, J.; et al. Rational design of biomimetic molecularly imprinted materials: Theoretical and computational strategies for guiding nanoscale structured polymer development. Anal. Bioanal. Chem 2011, 400, 1771–1786. [Google Scholar]
- Sajonz, P.; Kele, M.; Zhong, G.; Sellergren, B.; Guiochon, G. Study of the thermodynamics and mass transfer kinetics of two enantiomers on a polymeric imprinted stationary phase. J. Chromatogr. A 1998, 810, 1–17. [Google Scholar]
- Chen, Y.; Kele, M.; Sajonz, P.; Sellergren, B.; Guiochon, G. Influence of thermal annealing on the thermodynamic and mass-transfer kinetic properties of d- and l-Phenylalanine anilide on imprinted polymeric stationary phases. Anal. Chem 1999, 71, 928–938. [Google Scholar]
- Piletska, E.V.; Piletsky, S.A. Size matters: Influence of the size of nanoparticles on their interactions with ligands immobilized on the solid surface. Langmuir 2010, 26, 3783–3785. [Google Scholar]
- Karlsson, B.C.G.; Rosengren, A.M.; Näslund, I.; Andersson, P.O.; Nicholls, I.A. Synthetic human albumin Sudlow I binding site mimics. J. Med. Chem 2010, 53, 7932–7937. [Google Scholar]
- Li, T.; Chang, C.-Y.; Jin, D.-Y.; Lin, P.-J.; Khvorova, A.; Stafford, D.W. Identification of the gene for vitamin K epoxide reductase. Nature 2004, 427, 541–544. [Google Scholar]
- Rost, S.; Fregin, A.; Ivaskevicius, V.; Conzelmann, E.; Hortnagel, K.; Pelz, H.-J.; Lappegard, K.; Seifried, E.; Scharrer, I.; Tuddenham, E.G.D.; et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004, 427, 537–541. [Google Scholar]
- Karlsson, B.C.G.; Rosengren, A.M.; Andersson, P.O.; Nicholls, I.A. The spectrophysics of warfarin: Implications for protein binding. J. Phys. Chem. B 2007, 111, 10520–10528. [Google Scholar]
- Karlsson, B.C.G.; Rosengren, A.M.; Andersson, P.O.; Nicholls, I.A. Molecular insights on the two fluorescence lifetimes displayed by warfarin from fluorescence anisotropy and molecular dynamics studies. J. Phys. Chem. B 2009, 113, 7945–7949. [Google Scholar]
- Rosengren, A.M.; Karlsson, B.C.G.; Näslund, I.; Andersson, P.O.; Nicholls, I.A. In situ detection of warfarin using time-correlated single-photon counting. Biochem. Bioph. Res. Commun 2011, 407, 60–62. [Google Scholar]
- Rosengren, A.M.; Karlsson, B.C.G.; Nicholls, I.A. Monitoring the distribution of warfarin in blood plasma. ACS Med. Chem. Lett 2012, 3, 650–652. [Google Scholar]
- He, M.; Korzekwa, K.R.; Jones, J.P.; Rettie, A.E.; Trager, W.F. Structural forms of phenprocoumon and warfarin that are metabolized at the active site of CYP2C9. Arch. Biochem. Biophys 1999, 372, 16–28. [Google Scholar]
- Valente, E.J.; Lingafelter, E.C.; Porter, W.R.; Trager, W.F. Structure of warfarin in solution. J. Med. Chem 1977, 20, 1489–1493. [Google Scholar]
- Nicholls, I.A.; Karlsson, B.C.G.; Rosengren, A.M.; Henschel, H. Warfarin: An environment-dependent switchable molecular probe. J. Mol. Recognit 2010, 23, 604–608. [Google Scholar]
- Rosengren, A.M.; Karlsson, B.C.G. Spectroscopic evidence for the presence of the cyclic hemiketal form of warfarin in aqeous solution: Consequences for bioavailability. Biochem. Biophys. Res. Commun 2011, 407, 318–320. [Google Scholar]
- Lange, H. Determination of the degree of swelling and cross-linking of extremely small polymer gel quantities by analytical ultracentrifugation. Colloid Polym. Sci 1986, 264, 488–493. [Google Scholar]
- Brunauer, S.; Emmett, P.H.; Teller, E. Adsorption of gases in multimolecular layers. J. Am. Chem. Soc 1938, 60, 309–319. [Google Scholar]
- Langmuir, I. The constitution and fundamental properties of solids and liquids. Part I. Solids. J. Am. Chem. Soc 1916, 38, 2221–2295. [Google Scholar]
- Gregg, S.J.; Sing, K.S.W. Adsorption, Surface and Porosity, 2nd ed; Academic Press: London, UK, 1982. [Google Scholar]
- Barrett, E.P.; Joyner, L.G.; Halenda, P.P. The determination of pore volume and area distributions in porous substances. I. Computations from nitrogen isotherms. J. Am. Chem. Soc 1951, 73, 373–380. [Google Scholar]


| Components | MIP1 | MIP2 | MIP3 | MIP4 | MIP5 | REF1 | REF2 | REF3 | REF4 | REF5 |
|---|---|---|---|---|---|---|---|---|---|---|
| Warfarin (mmol) | 1.39 | 1.39 | 1.39 | 1.39 | 1.39 | |||||
| MAA (mmol) | 2.78 | 6.95 | 16.68 | 27.80 | 55.60 | 2.78 | 6.95 | 16.68 | 27.80 | 55.60 |
| EDMA (mmol) | 76.5 | 76.5 | 76.5 | 76.5 | 76.5 | 76.5 | 76.5 | 76.5 | 76.5 | 76.45 |
| CHCl3 (mL) | 23.5 | 24.0 | 25.3 | 26.8 | 30.6 | 23.5 | 24.0 | 25.3 | 26.8 | 30.6 |
| AIBN (mmol) | 2.02 | 2.08 | 2.20 | 2.35 | 2.71 | 2.02 | 2.08 | 2.20 | 2.35 | 2.71 |
| EDMA:MAA ratio | 27.5 | 11.0 | 4.6 | 2.8 | 1.4 | 27.5 | 11.0 | 4.6 | 2.8 | 1.4 |
| Polymer | Average pore size (nm) | |
|---|---|---|
| peak 1 | peak 2 | |
| MIP3 | 2.1 | 3.9 |
| MIP4 | 3.6 | 22 |
| MIP5 | 3.4 | 100 |
| REF3 | 3.6 | 15 |
| REF4 | 3.6 | 37 |
| REF5 | 3.4 | 114 |
| Polymer | Surface area (m2/g) (r) a | PC50 (mg/mL) b | Sr c | |
|---|---|---|---|---|
| BET | Langmuir | |||
| MIP1 | 35.4 ± 1.8 [0.994] | 51.3 ± 2.2 [0.996] | 13.2 | 2.50 |
| MIP2 | 3.6 ± 0.5 [0.962] | 5.8 ± 1.3 [0.917] | 12.4 | 2.65 |
| MIP3 | 73.5 ± 1.2 [0.999] | 102.0 ± 3.1 [0.998] | 7.3 | 2.33 |
| MIP4 | 118.4 ± 0.6 [1.000] | 163.5 ± 4.3 [0.999] | 4.7 | 1.60 |
| MIP5 | 50.0 ± 0.2 [1.000] | 69.5 ± 2.3 [0.998] | 12.7 | 1.30 |
| REF1 | 2.3 ± 0.1 [0.999] | 3.0 ± 0.0 [1.000] | 13.5 | 2.40 |
| REF2 | 2.6 ± 0.1 [0.996] | 3.6 ± 0.2 [0.992] | 11.9 | 2.60 |
| REF3 | 238.3 ± 2.5 [1.000] | 329.9 ± 8.4 [0.999] | 9.0 | 2.00 |
| REF4 | 169.1 ± 1.2 [1.000] | 233.4 ± 5.7 [0.999] | 7.1 | 1.35 |
| REF5 | 83.3 ± 0.4 [1.000] | 116.1 ± 3.1 [0.999] | 9.9 | 1.30 |
© 2013 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Rosengren, A.M.; Karlsson, B.C.G.; Nicholls, I.A. Consequences of Morphology on Molecularly Imprinted Polymer-Ligand Recognition. Int. J. Mol. Sci. 2013, 14, 1207-1217. https://doi.org/10.3390/ijms14011207
Rosengren AM, Karlsson BCG, Nicholls IA. Consequences of Morphology on Molecularly Imprinted Polymer-Ligand Recognition. International Journal of Molecular Sciences. 2013; 14(1):1207-1217. https://doi.org/10.3390/ijms14011207
Chicago/Turabian StyleRosengren, Annika M., Björn C. G. Karlsson, and Ian A. Nicholls. 2013. "Consequences of Morphology on Molecularly Imprinted Polymer-Ligand Recognition" International Journal of Molecular Sciences 14, no. 1: 1207-1217. https://doi.org/10.3390/ijms14011207





