Regulation of MIR Genes in Response to Abiotic Stress in Hevea brasiliensis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Relative Accumulation of premiR in Response to Abiotic Stress in in Vitro Plantlets
2.2. Relative Accumulation of preMIR in Response to Ethylene (ET), Methyl Jasmonate (MeJA) and Wounding in Three-Month-Old Epicormic Shoots
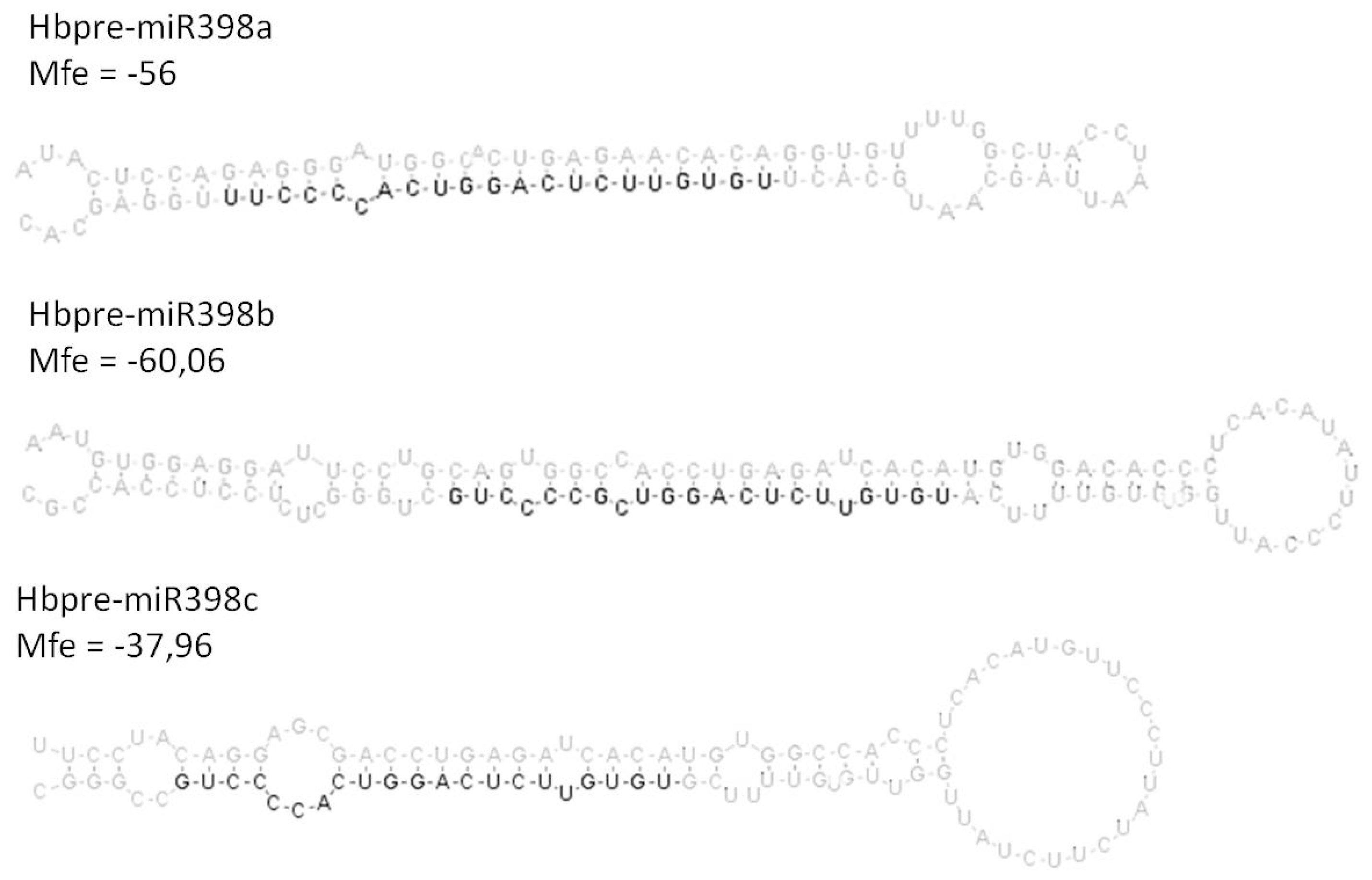
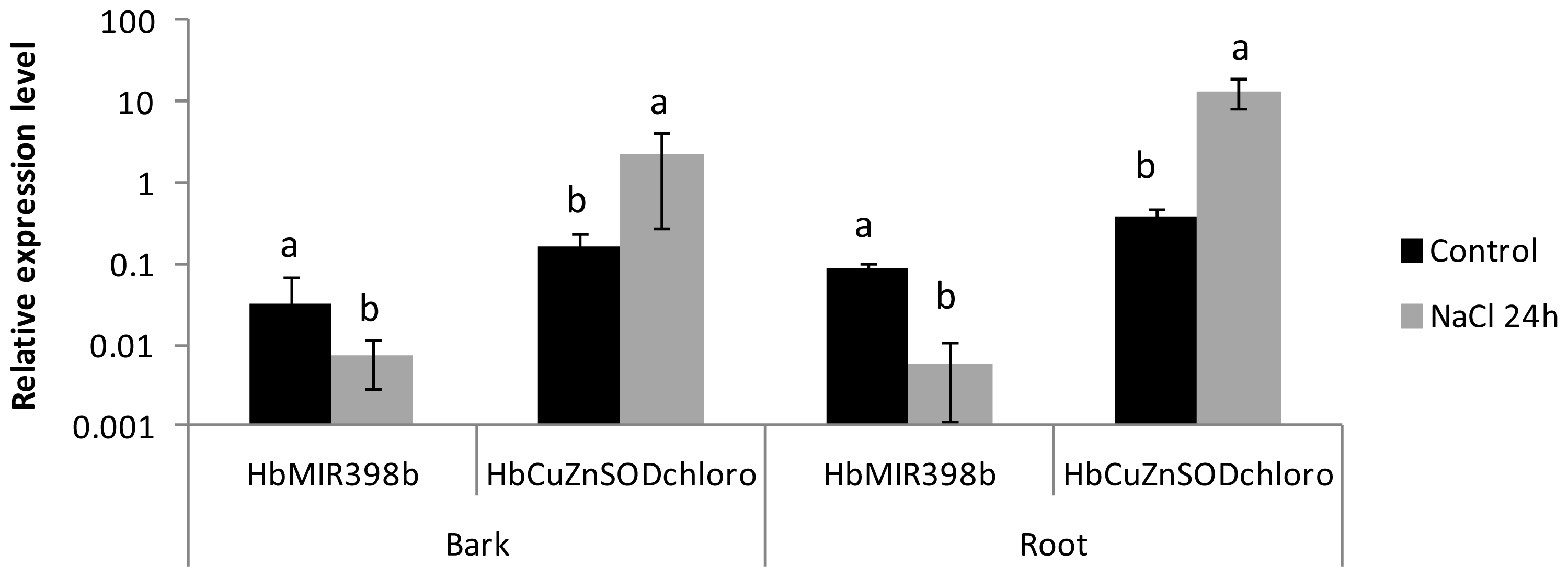
2.3. Analysis of the Co-Regulated Expression of the HbMIR398a, HbMIR398b and HbMIR398c Genes and Their Putative Target Gene Chloroplastic HbCuZnSOD in Response to Abiotic Stress and to Hormone Treatments
3. Experimental Section
3.1. Plant Material and Treatments
3.2. Extraction and Purification of Total RNAs
3.3. Checking for the Presence of Genomic DNA and DNAse Treatment
3.4. cDNA Synthesis and Real-Time PCR
3.5. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-14-19587-s001.pdf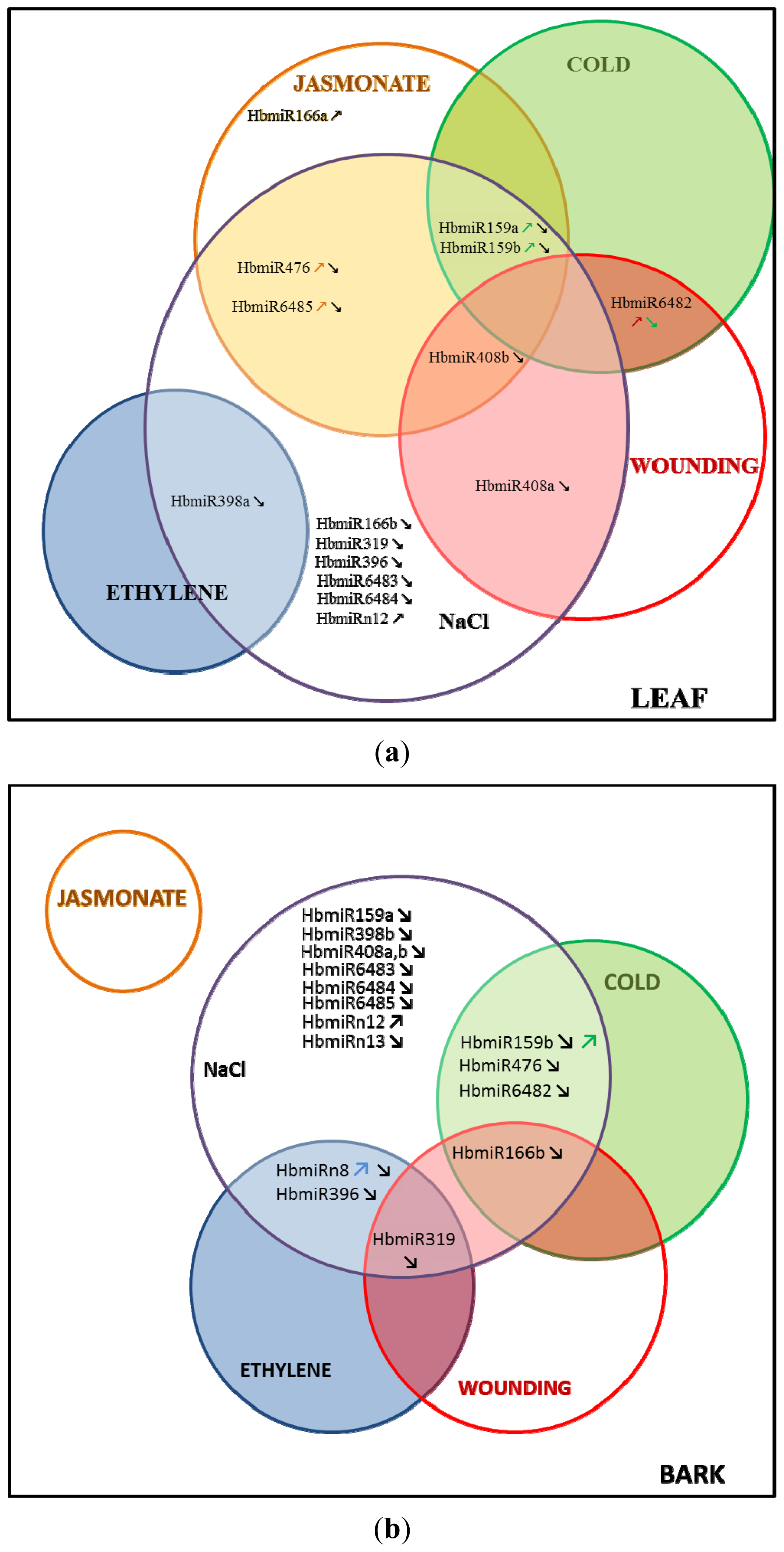
| Gene | Tissue | Expression value for control | Cold/C | p-value | NaCl/C | p-value |
|---|---|---|---|---|---|---|
| HbpremiR156 | leaf | 1.72 × 10−3a | 1.79 | 0.52 | 0.17 | 0.08 |
| bark | 1.80 × 10−3a | 0.91 | 0.72 | 0.20 | 0.08 | |
| root | 2.63 × 10−3a | 0.70 | 0.39 | 0.39 | 0.13 | |
| HbpremiR159a | leaf | 4.68 × 10−2b | 3.45 | 0.008 | 0.04 | 0.002 |
| bark | 7.45 × 10−2a | 1.25 | 0.22 | 0.17 | <0.001 | |
| root | 5.94 × 10−2a,b | 0.32 | 0.02 | 0.10 | 0.01 | |
| HbpremiR159b | leaf | 1.56 × 10−1a | 4.06 | 0.004 | 0.05 | 0.003 |
| bark | 2.21 × 10−1a | 1.59 | 0.03 | 0.23 | <0.0001 | |
| root | 1.94 × 10−1a | 0.44 | 0.23 | 0.11 | 0.02 | |
| HbpremiR166a | leaf | 1.19 × 10−2a | 1.38 | 0.38 | 2.08 | 0.08 |
| bark | 1.78 × 10−2a | 2.00 | 0.18 | 0.70 | 0.16 | |
| root | 4.16 × 10−2a | 0.60 | 0.62 | 2.76 | 0.13 | |
| HbpremiR166b | leaf | 2.21 × 10−1c | 1.34 | 0.31 | 0.20 | 0.01 |
| bark | 5.20 × 100a | 0.64 | 0.01 | 0.14 | 0.003 | |
| root | 9.67 × 10−1b | 0.48 | 0.39 | 1.01 | 0.80 | |
| HbpremiR319 | leaf | 5.73 × 10−3c | 0.29 | 0.08 | 0.23 | 0.02 |
| bark | 9.22 × 10−2a | 1.30 | 0.90 | 0.07 | <0.0001 | |
| root | 1.76 × 10−2b | nc | – | nc | – | |
| HbpremiR396 | leaf | 4.73 × 10−1a | 1.34 | 0.28 | 0.10 | 0.002 |
| bark | 1.41 × 100a | 0.93 | 0.61 | 0.17 | <0.0001 | |
| root | 2.33 × 100a | 0.28 | 0.19 | 0.12 | 0.05 | |
| HbpremiR398a | leaf | 6.93 × 10−3a | 0.34 | 0.12 | 0.21 | 0.02 |
| bark | 2.27 × 10−3a,b | 0.23 | 0.31 | 0.49 | 0.64 | |
| root | 6.78 × 10−4b | 0.47 | 0.39 | 0.10 | 0.20 | |
| HbpremiR398b | leaf | 3.27 × 10−2b | 0.95 | 0.92 | 0.87 | 0.61 |
| bark | 8.68 × 10−2a | 0.52 | 0.15 | 0.07 | 0.01 | |
| root | 3.19 × 10−2b | 0.62 | 0.34 | 0.15 | 0.03 | |
| HbpremiR398c | leaf | trace | nc | – | nc | – |
| bark | trace | nc | – | nc | – | |
| root | trace | nc | – | nc | – | |
| HbpremiR408a | leaf | 8.47 × 10−1a | 0.47 | 0.20 | 0.10 | <0.0001 |
| bark | 1.50 × 100a | 0.23 | 0.11 | 0.07 | <0.001 | |
| root | 1.48 × 100a | 0.22 | 0.02 | 0.06 | 0.01 | |
| HbpremiR408b | leaf | 1.60 × 100b | 0.48 | 0.21 | 0.12 | <0.0001 |
| bark | 3.54 × 100a | 0.22 | 0.11 | 0.08 | 0.01 | |
| root | 2.75 × 100a,b | 0.22 | 0.01 | 0.08 | 0.001 | |
| HbpremiR476 | leaf | 2.71 × 10−2a | 0.66 | 0.39 | 0.16 | 0.01 |
| bark | 2.84 × 10−2a | 0.36 | 0.01 | 0.37 | 0.002 | |
| root | 8.49 × 10−2a | 0.20 | 0.11 | 0.41 | 0.23 | |
| HbpremiR6482 | leaf | 2.20 × 100c | 0.09 | 0.001 | 0.30 | 0.06 |
| bark | 9.86 × 100b | 0.21 | 0.002 | 0.45 | 0.01 | |
| root | 7.06 × 10a | 0.05 | 0.004 | 0.05 | 0.003 | |
| HbpremiR6483 | leaf | 5.47 × 10a | 0.59 | 0.50 | 0.03 | 0.002 |
| bark | 1.56 × 10a | 0.70 | 0.43 | 0.18 | 0.01 | |
| root | 5.54 × 10−1b | 1.05 | 0.72 | 0.51 | 0.59 | |
| HbpremiR6484 | leaf | 7.17 × 10−1a | 0.70 | 0.26 | 0.22 | 0.003 |
| bark | 7.47 × 10−1a | 0.65 | 0.06 | 0.55 | 0.02 | |
| root | 3.47 × 10−1b | 0.25 | 0.02 | 0.35 | 0.05 | |
| HbpremiR6485 | leaf | 7.44 × 10a | 0.73 | 0.49 | 0.03 | 0.000 |
| bark | 3.14 × 10a | 0.82 | 0.77 | 0.22 | 0.02 | |
| root | 6.06 × 100b | 0.73 | 0.14 | 0.27 | 0.01 | |
| HbpremiRn11 | leaf | trace | nc | – | nc | – |
| bark | trace | nc | – | nc | – | |
| root | trace | nc | – | nc | – | |
| HbpremiRn12 | leaf | 7.26 × 10−2a | 1.25 | 0.50 | 6.45 | <0.0001 |
| bark | 8.80 × 10−2a | 0.75 | 0.29 | 5.93 | <0.001 | |
| root | 1.47 × 10−1a | 0.48 | 0.10 | 4.17 | 0.01 | |
| HbpremiRn13 | leaf | 3.77 × 102a | 1.17 | 0.60 | 0.34 | 0.11 |
| bark | 3.41 × 102a | 1.17 | 0.74 | 0.46 | 0.001 | |
| root | 2.65 × 102a | 0.81 | 0.51 | 1.90 | 0.06 | |
| Gene | Tissue | Expression valuefor control | ET/C | p-value | MeJA/C | p-value | W/C | p-value |
|---|---|---|---|---|---|---|---|---|
| HbpremiR156 | leaves | 4.42 × 10−4a | 1.02 | 0.94 | 6.23 | 0.06 | 2.92 | 0.68 |
| bark | 6.80 × 10−4a | 1.63 | 0.26 | 0.66 | 0.62 | 1.68 | 0.35 | |
| HbpremiR159a | leaves | 2.34 × 10−1a | 1.22 | 0.91 | 1.40 | 0.04 | 1.04 | 0.88 |
| bark | 4.41 × 10−2b | 0.40 | 0.30 | 0.75 | 0.99 | 0.33 | 0.20 | |
| HbpremiR159b | leaves | 6.28 × 10−1a | 0.81 | 0.38 | 1.64 | 0.02 | 0.76 | 0.14 |
| bark | 1.96 × 10−1b | 0.50 | 0.35 | 0.86 | 0.92 | 0.41 | 0.22 | |
| HbpremiR166a | leaves | 4.02 × 10−2a | 0.49 | 0.39 | 2.84 | 0.05 | 0.98 | 0.84 |
| bark | 1.62 × 10−2a | 0.35 | 0.17 | 1.00 | 0.99 | 1.06 | 0.72 | |
| HbpremiR166b | leaves | 5.98 × 100a | 1.50 | 0.24 | 2.45 | 0.22 | 1.78 | 0.07 |
| bark | 1.00 × 10−1b | 1.08 | 0.67 | 1.81 | 0.15 | 0.32 | 0.03 | |
| HbpremiR319 | leaves | 2.80 × 10−1a | 0.73 | 0.63 | 1.46 | 0.33 | 0.87 | 0.83 |
| bark | 1.15 × 10−2b | 0.31 | 0.03 | 1.28 | 0.41 | 0.18 | 0.02 | |
| HbpremiR396 | leaves | 2.87 × 100a | 0.62 | 0.20 | 3.19 | 0.09 | 0.57 | 0.22 |
| bark | 4.80 × 10−1b | 0.27 | 0.02 | 0.50 | 0.13 | 0.43 | 0.07 | |
| HbpremiR398a | leaves | 3.61 × 10−4b | 0.22 | 0.05 | 0.69 | 0.53 | 1.85 | 0.34 |
| bark | 1.66 × 10−2a | 3.59 | 0.08 | 3.38 | 0.21 | 2.99 | 0.19 | |
| HbpremiR398b | leaves | 1.89 × 10−3b | 0.52 | 0.67 | 0.09 | 0.41 | 0.05 | 0.27 |
| bark | 5.01 × 10−2a,b | 15.19 | 0.09 | 0.61 | 0.46 | 4.66 | 0.60 | |
| HbpremiR398c | leaves | trace | 0.91 | 0.82 | nc | – | 0.04 | 0.16 |
| bark | trace | nc | – | nc | – | nc | – | |
| HbpremiR408a | leaves | 3.49 × 100a | 0.40 | 0.61 | 0.05 | 0.21 | 0.04 | 0.002 |
| bark | 1.29 × 100a | 0.50 | 0.64 | 0.002 | 0.52 | 0.05 | 0.62 | |
| HbpremiR408b | leaves | 5.40 × 100a | 0.42 | 0.55 | 0.02 | 0.01 | 0.04 | 0.001 |
| bark | 2.52 × 100a | 0.73 | 0.30 | 0.003 | 0.30 | 0.05 | 0.91 | |
| HbpremiR476 | leaves | 1.49 × 10−1a | 0.84 | 0.52 | 17.93 | 0.001 | 0.98 | 0.73 |
| bark | 1.73 × 10−2a | 0.26 | 0.10 | 6.18 | 0.06 | 0.32 | 0.30 | |
| HbpremiR6482 | leaves | 4.41 × 100a | 1.14 | 0.60 | 1.60 | 0.26 | 7.19 | 0.05 |
| bark | 1.63 × 100a | 0.62 | 0.48 | 1.32 | 0.53 | 2.68 | 0.14 | |
| HbpremiR6483 | leaves | 5.56 × 100a | 2.97 | 0.18 | 4.14 | 0.13 | 0.81 | 0.85 |
| bark | 8.63 × 100a | 1.13 | 0.67 | 1.34 | 0.69 | 0.68 | 0.53 | |
| HbpremiR6484 | leaves | 6.10 × 10−1a | 0.92 | 0.65 | 1.67 | 0.14 | 0.48 | 0.12 |
| bark | 8.92 × 10−1a | 0.99 | 1.00 | 1.15 | 0.72 | 0.73 | 0.41 | |
| HbpremiR6485 | leaves | 1.60 × 10b | 1.19 | 0.58 | 2.62 | 0.004 | 0.61 | 0.11 |
| bark | 3.00 × 10a | 1.00 | 0.94 | 0.82 | 0.44 | 0.52 | 0.06 | |
| HbpremiRn11 | leaves | trace | nc | – | 3.52 | 0.08 | 1.35 | 0.98 |
| bark | trace | 0.97 | 0.93 | 0.56 | 0.24 | 0.60 | 0.39 | |
| HbpremiRn12 | leaves | 6.19 × 10−2a | 1.23 | 0.65 | 1.51 | 0.12 | 7.12 | 0.13 |
| bark | 6.13 × 10−2a | 1.20 | 0.58 | 0.84 | 0.96 | 1.42 | 0.44 | |
| HbpremiRn13 | leaves | 3.16 × 102a | 1.23 | 0.39 | 1.24 | 0.55 | 1.41 | 0.96 |
| bark | 3.92 × 102a | 1.85 | 0.16 | 0.91 | 0.55 | 1.85 | 0.63 | |
| Pre-microRNA | Forward primer | Reverse primer | PCR efficiency |
|---|---|---|---|
| Hbpre-miR156 | TGGTGATGTTGTTGACAGAAGATAGAGAGC | GCACAAAGGAGTGAGATGCAGAGTCC | 1.79 |
| Hbpre-miR159a | GGTTAAGAAGTGGAGCTCCTTGAAGTC | GCTCCCTTCAATCCAAACAAGGATC | 1.958 |
| Hbpre-miR159b | GTGGAGCTCCTTGAAGTCCAATAGAGG | AGAGCTCCCTTCAATCCAAACAAGG | 1.881 |
| Hbpre-miR166a | TTCTTTTTGAGGGGAATGTTGTCTGG | GGAATGAAGCCTGGTCCGAGGAG | 1.820 |
| Hbpre-miR166b | GGGGAATGTTGTCTGGTTCGATG | TCAAATCAAACCCTGTTGGGGG | 1.738 |
| Hbpre-miR319 | CCAGTCACGGTGGGCAATGGG | GGAGCTCCCTTCAGTCCAAGTACAGG | 1.847 |
| Hbpre-miR396 | TGACCCTCTTCGTATTCTTCCACAGC | CCCACAGCTTTATTGAACCGCAAC | 1.782 |
| Hbpre-mir398a | TGAGAACACAGGTGTTTTGGCTACC | GTGCTCCAAAGGGGTGACCTGAG | 1.879 |
| Hbpre-mir398b | ACCTGAGATCACATGTGGACACCC | GCGGTGGAGGAGAGCCCAG | 1.939 |
| Hbpre-mir398c | TGGCCACCCTCACATGTTCCC | CCGGCAGGGGTGACCTGAG | 1.965 |
| Hbpre-miR408a | ACTGGGAACAGGCAGAGCATGG | GCCACAAGCCAGGGAAGAGGC | 1.723 |
| Hbpre-miR408b | GACATACAAAGACTGGGAACAGGCAG | GCCACAAGCCAGGGAAGAGGC | 1.792 |
| Hbpre-miR476 | GCCTTGTATGTTTCATTTAGTAATCCTTCT | GATAATCCTTCTATGCAAAGTCTTTTATGC | 1.732 |
| Hbpre-miR6482 | ACCAGGAACTGGTATCAACCCAGC | TGCTACCAATGAATCGGACCCACC | 1.837 |
| Hbpre-miR6483 | CAGTAAATAGCAGTATCGTGGATAGGG | GTCCAATCATTGATCCTGAAAATTTCTAC | 1.828 |
| Hbpre-miR6484 | TGGATTGGAGCCCAATACTGTGAC | CTGCTCCATTGATTTTACCATCTATGC | 1.873 |
| Hbpre-miR6485 | ACCTAGGATGTAGAAGAGCATAAC | ACTACATGAGTGGATATATAGGAATCC | 1.787 |
| Hbpre-miRn11 | GTATCAACGCAGATGTGCCGCC | CCCCAGCCAAACTCCCCACC | 1.828 |
| Hbpre-miRn12 | AGCTTTCACCCAATAACCTTTGCAGT | GCTCTTCCAATTCCTATCCAAAGTGGT | 1.78 |
| Hbpre-miRn13 | TGTGTTGGCCTTCGGGATCGG | CGAATGCCCCCGACTGTCCC | 1.889 |
| Hb-RH2b | GAGGTGGATTGGCTAACTGAGAAG | GTTGAACATCAAGTCCCCGAGC | 1.68 |
| HbCuZnSOD (flanking miRNA site) | GCTCTATCTCTCGCCGCCGCCTCC | CCGCAATTGTTGCTTCTGCC | 1.785 |
| HbCuZnSOD (3′UTR) | TGGCAGAAGCAACAATTGCGG | GCAGGGAACAATGGCTGCC | 2 |
| Treatment | Tissue | Ratio of relative transcript accumulation (Treated/Control) | ||||
|---|---|---|---|---|---|---|
| chloro CuZnSOD (flanking miRNA site) | chloro CuZnSOD (3′UTR) | preMIR398a | preMIR398b | preMIR398c | ||
| Cold | leaf | 2.71 | 3.76 | 0.34 | 0.95 | nc |
| bark | 8.98 | 8.90 | 0.23 | 0.52 | nc | |
| root | 3.41 | 1.46 | 0.47 | 0.62 | nc | |
| NaCl | leaf | 0.84 | 0.29 | 0.21 | 0.87 | nc |
| bark | 13.99 | 5.21 | 0.49 | 0.07 | nc | |
| root | 37.45 | 17.83 | 0.1 | 0.15 | nc | |
| Ethylene | leaf | nc | 0.42 | 0.22 | 0.52 | 0.91 |
| bark | 0.49 | 0.32 | 3.59 | 15.19 | nc | |
| MeJA | leaf | 1.47 | nc | 0.69 | 0.09 | nc |
| bark | 0.79 | 1.00 | 3.38 | 0.61 | nc | |
| Wounding | leaf | 2.61 | 7.07 | 1.85 | 0.05 | 0.04 |
| bark | 0.85 | 0.88 | 2.99 | 4.66 | nc | |
Acknowledgments
Conflicts of Interest
References
- Chen, S.; Peng, S.; Huang, G.; Wu, K.; Fu, X.; Chen, Z. Association of decreased expression of a Myb transcription factor with the TPD (tapping panel dryness) syndrome in Hevea brasiliensis. Plant Mol. Biol 2003, 51, 51–58. [Google Scholar]
- Strassner, J.; Schaller, F.; Frick, U.B.; Howe, G.A.; Weiler, E.W.; Amrhein, N.; Macheroux, P.; Schaller, A. Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J 2002, 32, 585–601. [Google Scholar]
- Orozco-Cardenas, M.L.; Narvaez-Vasquez, J.; Ryan, C.A. Hydrogen peroxide acts as a second messenger for the induction of defense genes in tomato plants in response to wounding, systemin, and methyl jasmonate. Plant Cell 2001, 13, 179–191. [Google Scholar]
- Chrestin, H. La stimulation à l’éthrel de l’hévéa jusqu’où ne pas aller trop loin. Caoutchouc et Plastiques 1985, 1985, 75–78. [Google Scholar]
- Faÿ, D. Histo- and cytopathology of trunk phloem necrosis, a form of rubber tree (Hevea brasiliensis Müll. Arg.) tapping panel dryness. Aust. J. Bot 2011, 59, 563–574. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Chen, X. MicroRNA biogenesis and function in plants. FEBS Lett 2005, 579, 5923–5931. [Google Scholar]
- Bonnet, E.; van de Peer, Y.; Rouze, P. The small RNA world of plants. New Phytol 2006, 171, 451–468. [Google Scholar]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. Plant microRNA: A small regulatory molecule with big impact. Dev. Biol 2006, 289, 3–16. [Google Scholar]
- Naqvi, A.; Sarwat, M.; Hasan, S.; Choudhury, N. Biogenesis, functions and fate of plant microRNAs. J. Cell. Physiol 2012, 227, 3163–3168. [Google Scholar]
- Mallory, A.C.; Bouche, N. MicroRNA-directed regulation: To cleave or not to cleave. Trends Plant Sci 2008, 13, 359–367. [Google Scholar]
- Mallory, A.C.; Elmayan, T.; Vaucheret, H. MicroRNA maturation and action—The expanding roles of ARGONAUTEs. Curr. Opin. Plant Biol 2008, 11, 560–566. [Google Scholar]
- Baulcombe, D. RNA silencing in plants. Kent 2004, 431, 356–363. [Google Scholar]
- Wu, L.; Zhou, H.; Zhang, Q.; Zhang, J.; Ni, F.; Liu, C.; Qi, Y. DNA methylation mediated by a microRNA pathway. Mol. Cell 2010, 38, 465–475. [Google Scholar]
- Chellappan, P.; Xia, J.; Zhou, X.; Gao, S.; Zhang, X.; Coutino, G.; Vazquez, F.; Zhang, W.; Jin, H. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 2010, 38, 6883–6894. [Google Scholar]
- Khraiwesh, B.; Zhu, J.K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar]
- Rhoades, M.W.; Reinhart, B.J.; Lim, L.P.; Burge, C.B.; Bartel, B.; Bartel, D.P. Prediction of plant microRNA targets. Cell 2002, 110, 513–520. [Google Scholar]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol 1998, 49, 249–279. [Google Scholar]
- Dat, J.; Vandenabeele, S.; Vranova, E.; van Montagu, M.; Inze, D.; van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci 2000, 57, 779–795. [Google Scholar]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar]
- Scandalios, J.G. Oxidative stress: Molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res 2005, 38, 995–1014. [Google Scholar]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 2011, 39, D152–D157. [Google Scholar]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 2006, 34, D140–D144. [Google Scholar]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res 2008, 36, D154–D158. [Google Scholar]
- Gébelin, V.; Argout, X.; Engchuan, W.; Pitollat, B.; Duan, C.; Montoro, P.; Leclercq, J. Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol 2012, 12. [Google Scholar] [CrossRef]
- Lertpanyasampatha, M.; Gao, L.; Kongsawadworakul, P.; Viboonjun, U.; Chrestin, H.; Liu, R.; Chen, X.; Narangajavana, J. Genome-wide analysis of microRNAs in rubber tree (Hevea brasiliensis L.) using high-throughput sequencing. Planta 2012, 236, 437–445. [Google Scholar]
- Gébelin, V.; Leclercq, J.; Kuswanhadi Argout, X.; Chaidamsari, T.; Hu, S.; Tang, C.; Sarah, G.; Yang, M.; Montoro, P. The small RNA profile in latex from Hevea brasiliensis trees is affected by Tapping Panel Dryness. Tree Physiol 2013, in press.. [Google Scholar]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar]
- Duan, C.; Argout, X.; Gebelin, V.; Summo, M.; Dufayard, J.F.; Leclercq, J.; Hadi, K.; Piyatrakul, P.; Pirrello, J.; Rio, M.; et al. Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genomics 2013, 14. [Google Scholar] [CrossRef]
- Duan, C.; Rio, M.; Leclercq, J.; Bonnot, F.; Oliver, G.; Montoro, P. Gene expression pattern in response to wounding, methyl jasmonate and ethylene in the bark of Hevea brasiliensis. Tree Physiol 2010, 30, 1349–1359. [Google Scholar]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J 2007, 49, 592–606. [Google Scholar]
- Lu, S.; Sun, Y.H.; Shi, R.; Clark, C.; Li, L.; Chiang, V.L. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsi s. Plant Cell 2005, 17, 2186–2203. [Google Scholar]
- Sanan-Mishra, N.; Kumar, V.; Sopory, S.K.; Mukherjee, S.K. Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Mol. Genet. Genomics 2009, 282, 463–474. [Google Scholar]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar]
- Lu, S.; Sun, Y.H.; Chiang, V.L. Stress-responsive microRNAs in Populus. Plant J 2008, 55, 131–151. [Google Scholar]
- Liu, H.H.; Tian, X.; Li, Y.J.; Wu, C.A.; Zheng, C.C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 2008, 14, 836–843. [Google Scholar]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot 2009, 103, 29–38. [Google Scholar]
- Ben Chaabane, S.; Liu, R.; Chinnusamy, V.; Kwon, Y.; Park, J.-H.; Kim, S.Y.; Zhu, J.-K.; Yang, S.W.; Lee, B.-H. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res 2013, 41, 1984–1997. [Google Scholar]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar]
- Axtell, A. JPM patient information. Depression in palliative care. J. Palliat. Med 2008, 11, 529–530. [Google Scholar]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant 2011, 143, 1–9. [Google Scholar]
- Lardet, L.; Martin, F.; Dessailly, F.; Carron, M.P.; Montoro, P. Effect of exogenous calcium on post-thaw growth recovery and subsequent plant regeneration of cryopreserved embryogenic calli of Hevea brasiliensis (Mull. Arg.). Plant Cell Rep 2007, 26, 559–569. [Google Scholar]
- Piyatrakul, P.; Putranto, R.A.; Martin, F.; Rio, M.; Dessailly, F.; Leclercq, J.; Dufayard, J.F.; Lardet, L.; Montoro, P. Some ethylene biosynthesis and AP2/ERF genes reveal a specific pattern of expression during somatic embryogenesis in Hevea brasiliensis. BMC Plant Biol 2012, 12. [Google Scholar] [CrossRef]
- German, M.A.; Luo, S.; Schroth, G.; Meyers, B.C.; Green, P.J. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nat. Protoc 2009, 4, 356–362. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Gébelin, V.; Leclercq, J.; Hu, S.; Tang, C.; Montoro, P. Regulation of MIR Genes in Response to Abiotic Stress in Hevea brasiliensis. Int. J. Mol. Sci. 2013, 14, 19587-19604. https://doi.org/10.3390/ijms141019587
Gébelin V, Leclercq J, Hu S, Tang C, Montoro P. Regulation of MIR Genes in Response to Abiotic Stress in Hevea brasiliensis. International Journal of Molecular Sciences. 2013; 14(10):19587-19604. https://doi.org/10.3390/ijms141019587
Chicago/Turabian StyleGébelin, Virginie, Julie Leclercq, Songnian Hu, Chaorong Tang, and Pascal Montoro. 2013. "Regulation of MIR Genes in Response to Abiotic Stress in Hevea brasiliensis" International Journal of Molecular Sciences 14, no. 10: 19587-19604. https://doi.org/10.3390/ijms141019587
APA StyleGébelin, V., Leclercq, J., Hu, S., Tang, C., & Montoro, P. (2013). Regulation of MIR Genes in Response to Abiotic Stress in Hevea brasiliensis. International Journal of Molecular Sciences, 14(10), 19587-19604. https://doi.org/10.3390/ijms141019587





