Screening and Functional Analysis of the Peroxiredoxin Specifically Expressed in Bursaphelenchus xylophilus—The Causative Agent of Pine Wilt Disease
Abstract
:1. Introduction
2. Results and Discussion
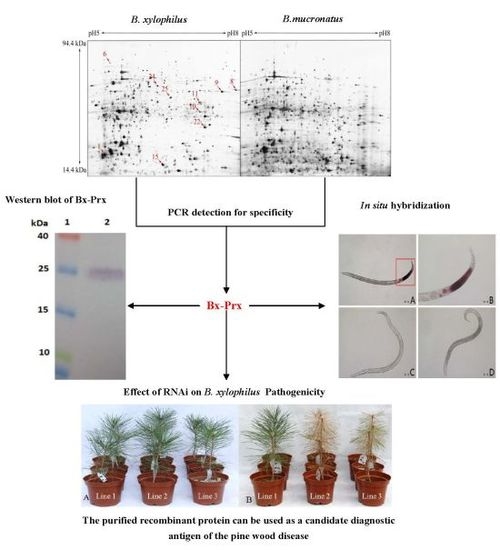
2.1. Comparative Proteome Analysis and Protein Identification
| Spot No. | Accession No. a | Protein Name | pI/kDa b | Score c | PN d | Molecular Function |
|---|---|---|---|---|---|---|
| 1 | BUX.s00713.141 | Actin | 42/5.3 | 264 | 6 | ATP binding, structural constituent of cytoskeleton |
| 6 | BUX.s00789.28 | Chaperonin Cpn60 | 60/5.39 | 213 | 13 | ATP binding |
| 8 | BUX_s01281.46 | GAPDH-1 | 37/7.68 | 419 | 10 | NAD+ activity |
| 9 | BUX.s01438.76 | Aldolase | 40/7.66 | 120 | 2 | fructose-bisphosphate aldolase activity |
| 10 | BUX.s01653.149 | Heat shock protein 70 | 71/5.55 | 528 | 14 | ATP binding |
| 11 | BUX.s01109.344 | Galectin | 33/6.22 | 146 | 5 | galactoside binding |
| 15 | BUX_s01226.18 | Cytosolic Fatty-acid binding | 88/6.14 | 71 | 9 | transporter activity |
| 24 | BUX.s00397.100 | Elongation factor 2 | 96/6.4 | 280 | 14 | translation elongation factor activity |
| 25 | BUX.s01143.143 | Aldo/keto reductase | 37/6.22 | 40 | 3 | oxidoreductase activity |
| 22 | BUX.s01109.415 | Peroxiredoxin | 22.1/6.09 | 156 | 6 | thioredoxin peroxidase activity |
2.2. PCR Detection for Specificity


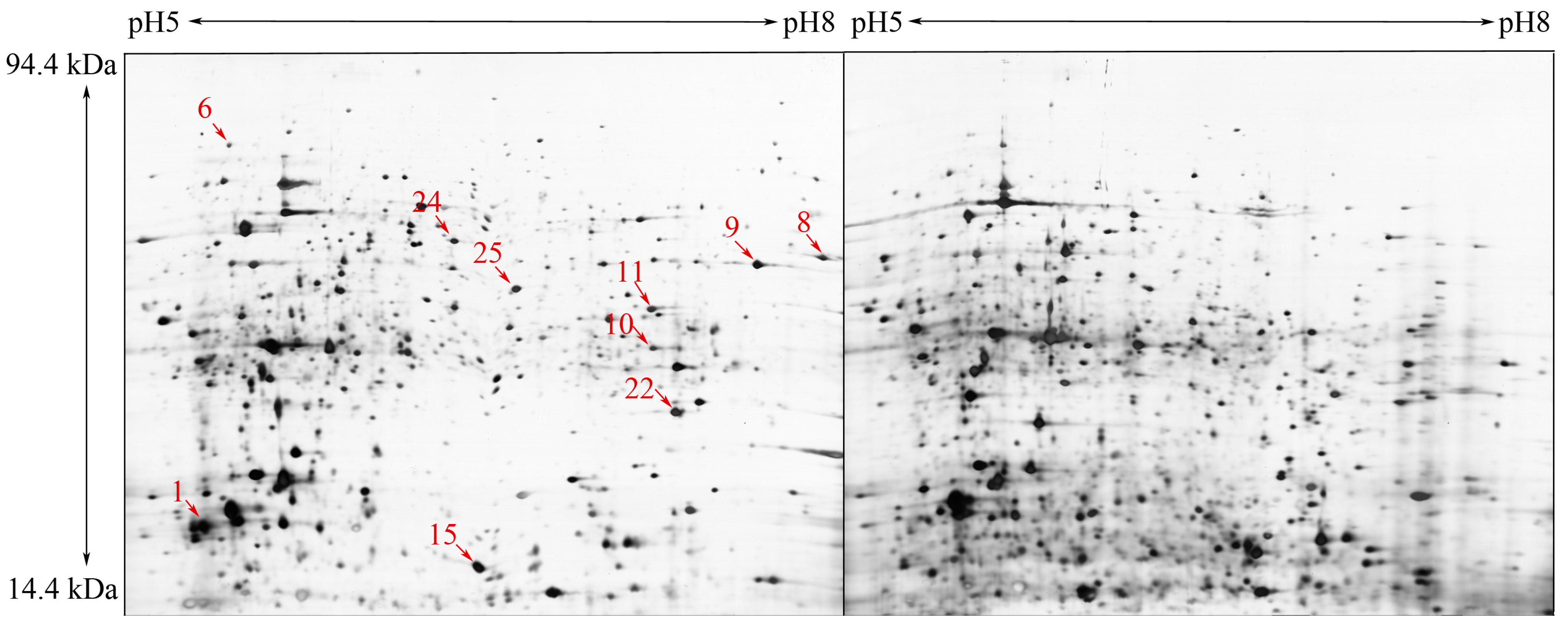
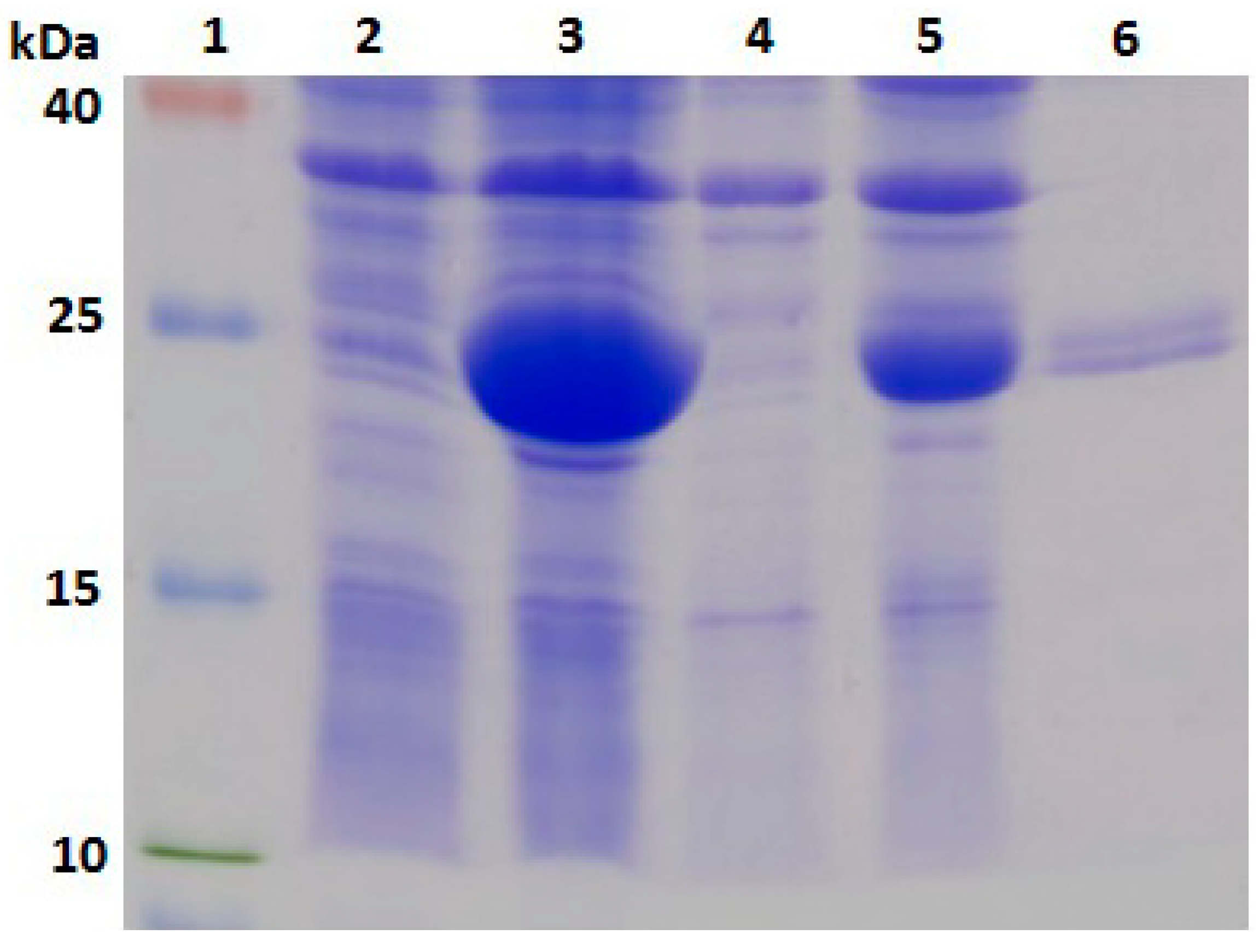
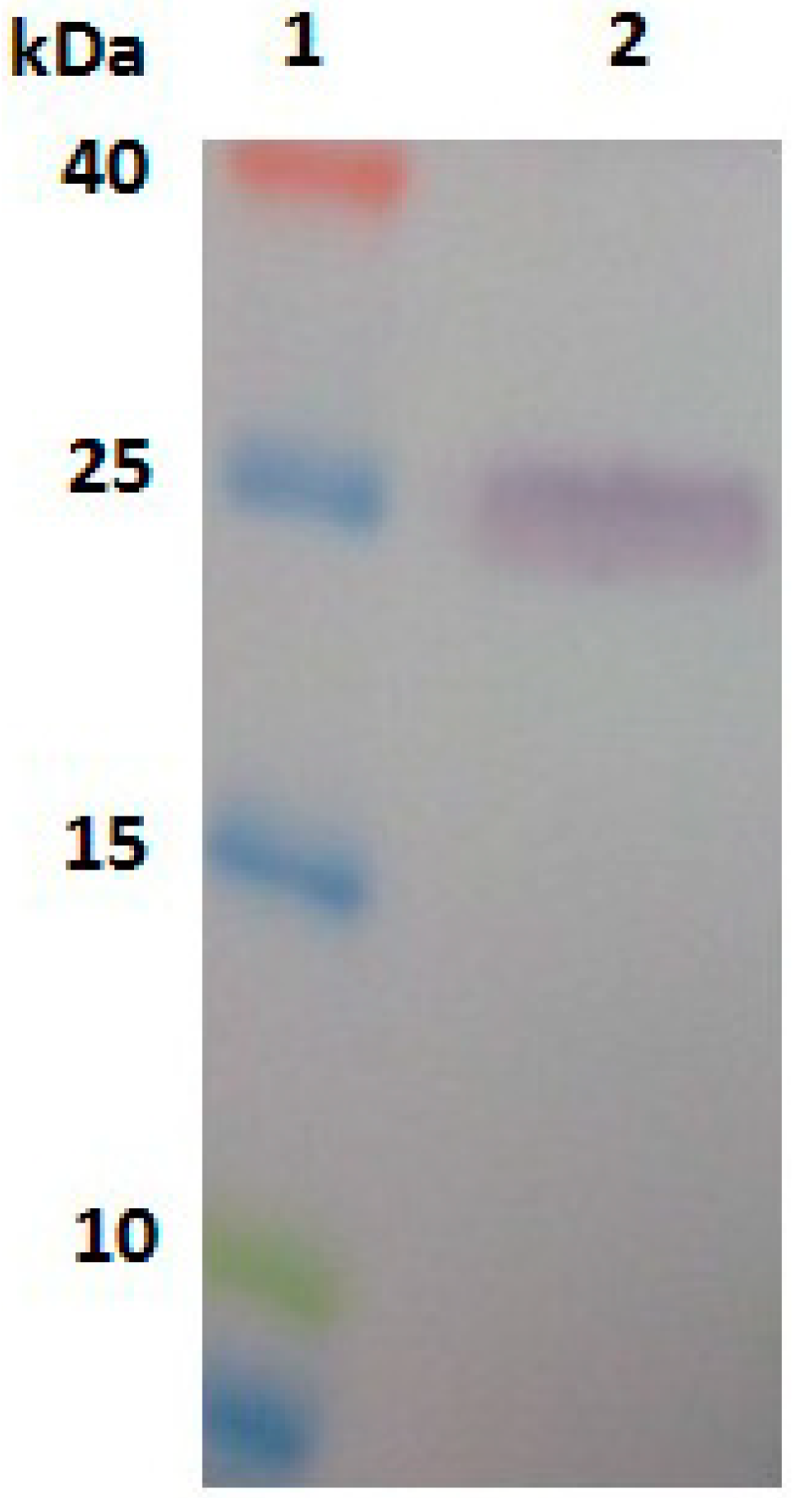
2.3. Expression, Purification and Identification of Bx-Prx
2.4. In Situ Hybridisation

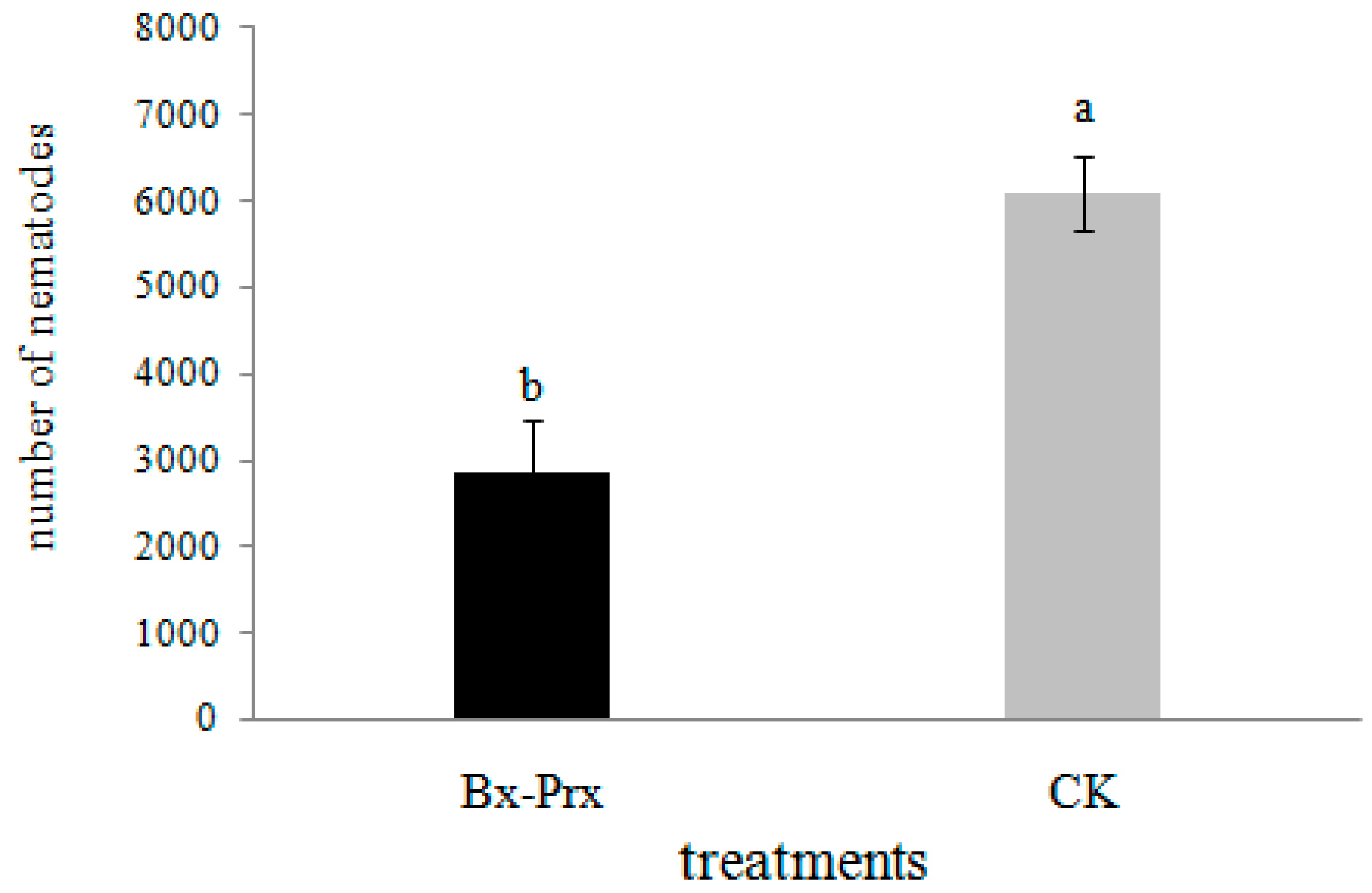
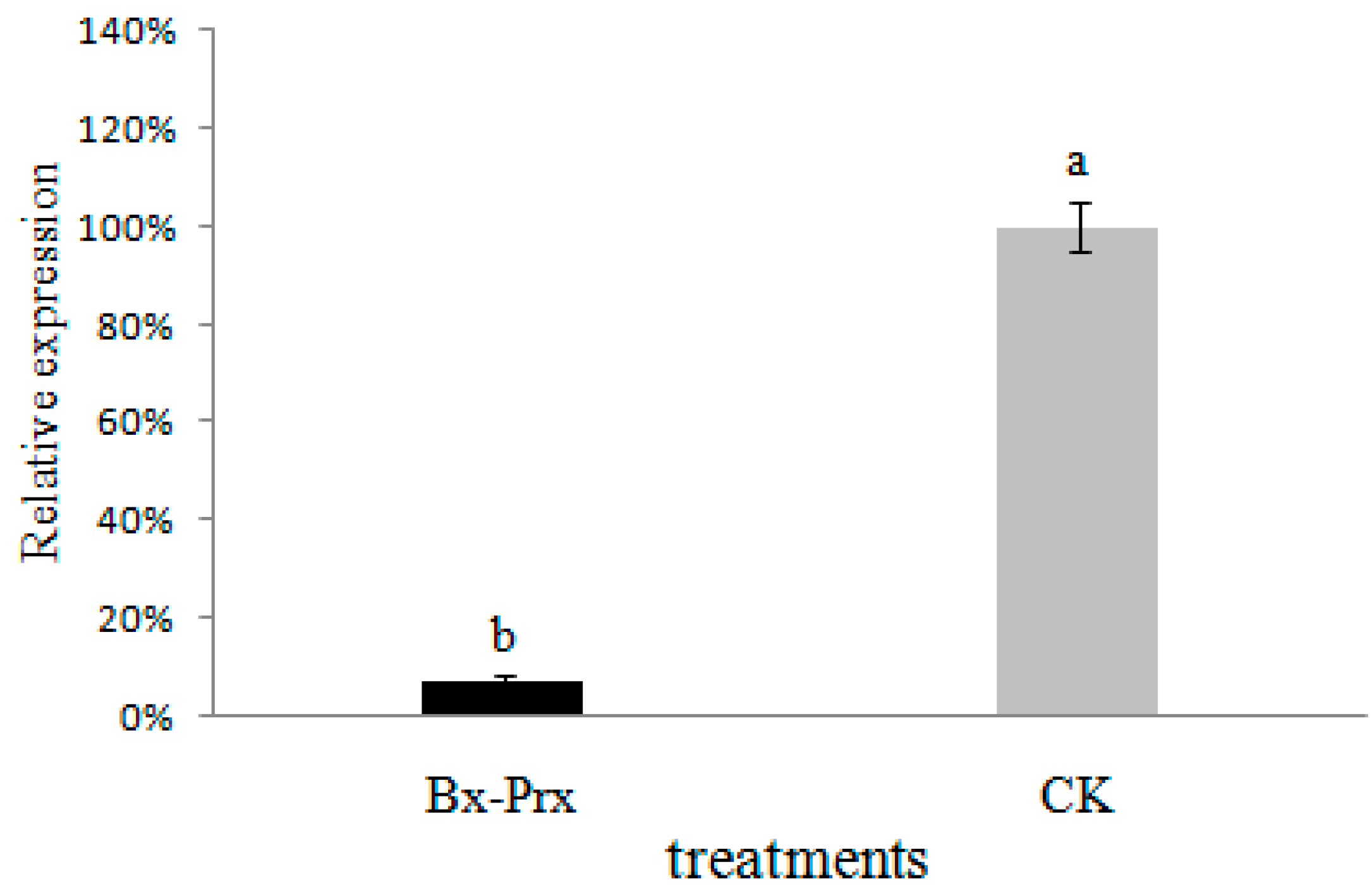
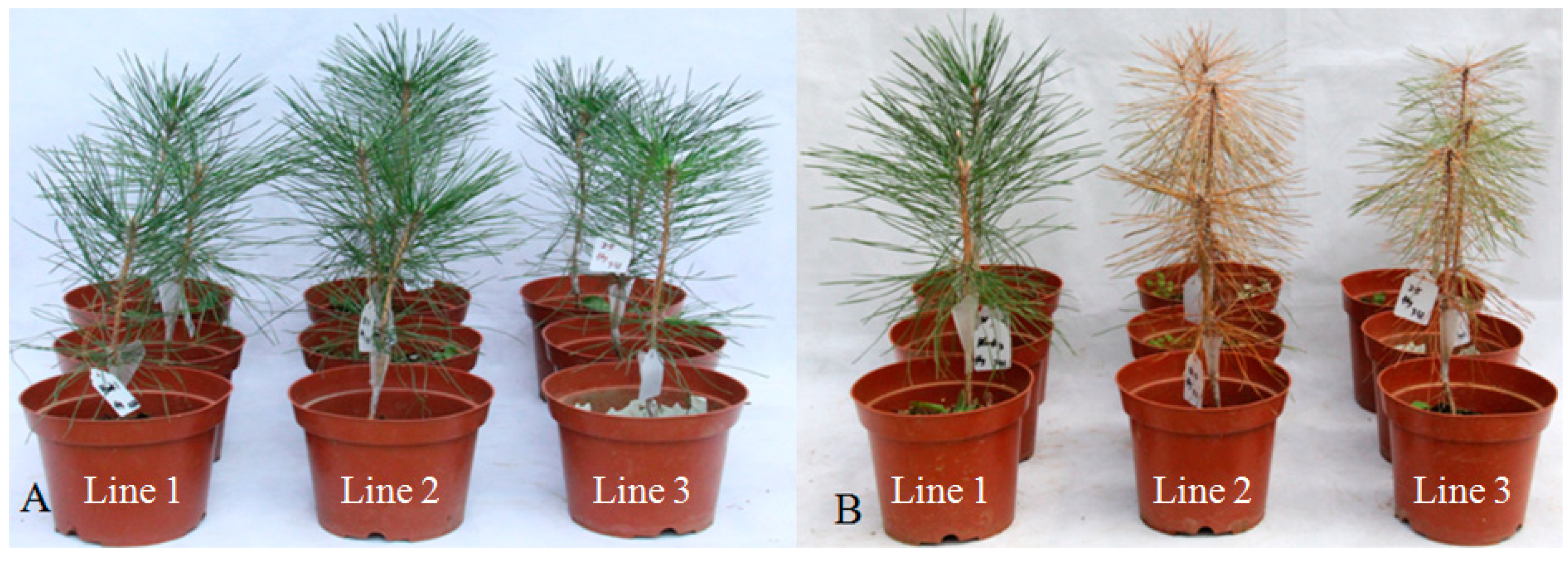
2.5. Effect of RNAi on B. xylophilus Reproduction and Pathogenicity
3. Experimental Section
3.1. Experimental Organisms
3.2. Protein Extraction
3.3. IEF and SDS PAGE
3.4. Image Analysis
3.5. PCR Detection for Specificity of Differential Proteins
| Gene | Spot No. | Sequence ID | Sense Primer (5-3) | Antisense Primer (5-3) |
|---|---|---|---|---|
| Actin-4 | 1 | gb|ACZ13341.1| | ATGTGTGACGAAGAAGTTGCCGCTC | TTAGAAACATTTGCGGTGAACGATG |
| Aldolase | 9 | gb|ACZ13345.1| | ATGGCCGAAGTCGGTGCTTCTTATC | TTAGTATGCGTGGTCTACGAGAGGG |
| Galectin-1 | 11 | gb|ACZ13331.1| | ATGACTGAGGAAAAGAAAACTTACA | TTAATGGATCTGGATGCCAGTGATC |
| Elongation factor 2 | 24 | gb|ACZ13348.1| | CCGGAATTCATGAATCCTTCGGTTTCACCGCTG | CCCAAGCTTTTAGAGTTTATCGTAGAAGTTGTCG |
| Bx-Prx | 22 | gb|ABW81468.1 | ATGTCCAAGGCTTTCATTGGCAAAC | TTAATGTTTGTTGAAATATTCGTGA |
3.6. Expression, Purification and Identification of Bx-Prx
3.7. In Situ Hybridisation
3.8. Bx-Prx Knockdown by RNA Interference (RNAi)
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jones, J.T.; Moens, M.; Mota, M.; Li, H.; Kikuchi, T. Bursaphelenchus xylophilus: Opportunities in comparative genomics and molecular host-parasite interactions. Mol. Plant Pathol. 2008, 9, 357–368. [Google Scholar] [CrossRef]
- Mamiya, Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 1983, 21, 201–220. [Google Scholar] [CrossRef]
- Mota, M.M.; Braasch, H.; Bravo, M.A.; Penas, A.C.; Burgermeister, W.; Metge, K.; Sousa, E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar] [CrossRef]
- Penas, A.C.; Correia, P.; Bravo, M.A.; Mota, M.; Tenreiro, R. Species of Bursaphelenchus Fuchs, 1937 (Nematoda: Parasitaphelenchidae) associated with maritime pine in Portugal. Nematology 2004, 6, 437–453. [Google Scholar] [CrossRef]
- Cao, A.; Liu, X.; Zhu, S.; Lu, B. Detection of the pinewood nematode, Bursaphelenchus xylophilus, using a real-time polymerase chain reaction assay. Phytopathology 2005, 95, 566–571. [Google Scholar] [CrossRef]
- Mamiya, Y.; Enda, N. Bursaphelenchus mucronatus n. sp. (Nematoda: Aphelenchoididae) from pine wood and its biology and pathogenicity to pine trees. Nematologica 1979, 25, 353–361. [Google Scholar] [CrossRef]
- Braasch, V.H. Pathogenitätstests mit Bursaphelenchus mucronatus an Kiefern und Fichtensämlingen in Deutschland. Eur. J. For. Pathol. 1996, 26, 205–216. [Google Scholar] [CrossRef]
- Tomminen, J. Pathogenicity studies with Bursaphelenchus mucronatus in Scots pine in Finland. Eur. J. For. Pathol. 1993, 23, 236–243. [Google Scholar] [CrossRef]
- Wingfield, M.; Blanchette, A.; Kondo, E. Comparison of the pine wood nematode, Bursaphelenchus xylophilus from pine and balsam fir1. Eur. J. For. Pathol. 1983, 13, 360–372. [Google Scholar] [CrossRef]
- Bolla, R.; Wood, R. Pathogenicity and potential for spread of the pine wood nematode Bursaphelenchus spp. In Sustainability of Pine Forests in Relation to Pine Wilt and Decline; Proceedings of International Symposium: Tokyo, Japan, 1998; pp. 3–13. [Google Scholar]
- Braasch, H.; Mota, M.; Vieira, P. Morphology of Bursaphelenchus xylophilus compared with other Bursaphelenchus species. In The Pinewood Nematode, Bursaphelenchus xylophilus, Proceedings of the International Workshop, Évora, Portugal, 20–22 August 2001; University of Évora: Évora, Portugal, August 2011. [Google Scholar]
- Cardoso, J.M.; Fonseca, L.; Abrantes, I. Direct molecular detection of the pinewood nematode, Bursaphelenchus xylophilus, from pine wood, bark and insect vector. Eur. J. Plant Pathol. 2012, 133, 419–425. [Google Scholar] [CrossRef]
- Bartosch, S.; Hartwig, C.; Spieck, E.; Bock, E. Immunological detection of nitrospira-like bacteria in various soils. Microb. Ecol. 2002, 43, 26–33. [Google Scholar] [CrossRef]
- Lin, N.; Hsu, Y.; Hsu, H. Immunological detection of plant viruses and a mycoplasmalike organism by direct tissue blotting on nitrocellulose membranes. Phytopathology 1990, 80, 824–828. [Google Scholar] [CrossRef]
- Lawler, C.; Joyce, P.; Harmey, M. Immunological differentiation between Bursaphelenchus xylophilus and B. Mucronatus. Nematologica 1993, 1, 536–546. [Google Scholar] [CrossRef]
- Burgermeister, W.; Metge, K.; Braasch, H.; Buchbach, E. Its-RFLP patterns for differentiation of 26 Bursaphelenchus species (Nematoda: Parasitaphelenchidae) and observations on their distribution. Rus. J. Nematol. 2005, 13, 29. [Google Scholar]
- Chen, F.M.; Negi, S.; Ye, J.R. A scar molecular marker to distinguish Bursaphelenchus mucronatus from the pinewood nematode, B. Xylophilus. For. Pathol. 2011, 41, 376–381. [Google Scholar] [CrossRef]
- Huang, L.; Ye, J.-R.; Wu, X.-Q.; Xu, X.-L.; Sheng, J.-M.; Zhou, Q.-X. Detection of the pine wood nematode using a real-time PCR assay to target the DNA topoisomerase I gene. Eur. J. Plant Pathol. 2010, 127, 89–98. [Google Scholar] [CrossRef]
- Irdani, T.; Marinari, A.; Bogani, P.; Ambrogioni, L.; Caroppo, S.; Buiatti, M. Molecular diversity among pine wood Bursaphelenchus populations detected by RAPD analysis. Redia 1995, 78, 149–161. [Google Scholar]
- Takemoto, S.; Kanzaki, N.; Futai, K. PCR-RFLP image analysis: A practical method for estimating isolate-specific allele frequency in a population consisting of two different strains of the pinewood nematode, Bursaphelenchus xylophilus (Aphelenchida: Aphelencoididae). Appl. Entomol. Zool. 2005, 40, 529–535. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Kanzaki, N.; Futai, K. Nematode, Bursaphelenchus xylophilus, from pine wood. Nematology 2005, 7, 775–782. [Google Scholar] [CrossRef]
- Zheng, J.; Subbotin, S.A.; He, S.; Gu, J.; Moens, M. Molecular characterisation of some Asian isolates of Bursaphelenchus xylophilus and B. Mucronatus using PCR-RFLPs and sequences of ribosomal DNA. Rus. J. Nematol. 2003, 11, 17–22. [Google Scholar]
- Zhuo, K.; Luo, M.; Cui, R.; Liao, J. A multiplex one-step PCR method for the simultaneous identification of Bursaphelenchus xylophilus, B. Mucronatus and B. Doui-three species within the xylophilus group. For. Pathol. 2011, 41, 66–69. [Google Scholar] [CrossRef]
- Kikuchi, T.; Aikawa, T.; Oeda, Y.; Karim, N.; Kanzaki, N. A rapid and precise diagnostic method for detecting the pinewood nematode Bursaphelenchus xylophilus by loop-mediated isothermal amplification. Phytopathology 2009, 99, 1365–1369. [Google Scholar] [CrossRef]
- Bogaerts, A.; Temmerman, L.; Boerjan, B.; Husson, S.J.; Schoofs, L.; Verleyen, P. A differential proteomics study of Caenorhabditis elegans infected with Aeromonas hydrophila. Dev. Comp. Immunol. 2010, 34, 690–698. [Google Scholar] [CrossRef]
- Jiang, D.; Malone, J.; Townsend, R.; Weil, G.J.; Li, B. Multiplex proteomics analysis of gender-associated proteins in Brugia malayi. Int. J. Parasitol. 2012, 42, 841–850. [Google Scholar]
- Shinya, R.; Morisaka, H.; Takeuchi, Y.; Ueda, M.; Futai, K. Comparison of the surface coat proteins of the pine wood nematode appeared during host pine infection and in vitro culture by a proteomic approach. Phytopathology 2010, 100, 1289–1297. [Google Scholar] [CrossRef]
- Shinya, R.; Morisaka, H.; Kikuchi, T.; Takeuchi, Y.; Ueda, M.; Futai, K. Secretome analysis of the pine wood nematode Bursaphelenchus xylophilus reveals the tangled roots of parasitism and its potential for molecular mimicry. PLoS One 2013, 8, e67377. [Google Scholar]
- Seo, H.-S.; Hirano, M.; Shibato, J.; Rakwal, R.; Hwang, I.K.; Masuo, Y. Effects of coffee bean aroma on the rat brain stressed by sleep deprivation: A selected transcript-and 2D gel-based proteome analysis. J. Agric. Food Chem. 2008, 56, 4665–4673. [Google Scholar] [CrossRef]
- Fang, X.P.; Ma, H.S.; Lu, D.Z.; Yu, H.; Lai, W.; Ruan, S. Comparative proteomics analysis of proteins expressed in the I-1 and I-2 internodes of strawberry stolons. Prot. Sci. 2011, 9, 26–41. [Google Scholar]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Ni, F.; Wang, Y.; Zhang, J.; Yu, L.; Fang, W.; Luo, D. Cathepsin B-like and hemoglobin-type cysteine proteases: Stage-specific gene expression in Angiostrongy cantonensis. Exp. Parasitol. 2012, 131, 433–441. [Google Scholar] [CrossRef]
- Li, Z.; Liu, X.; Chu, Y.; Wang, Y.; Zhang, Q.; Zhou, X. Cloning and characterization of a 2-Cys peroxiredoxin in the pine wood nematode, Bursaphelenchus xylophilus, a putative genetic factor facilitating the infestation. Int. J. Biol. Sci. 2011, 7, 823. [Google Scholar]
- Hudson, A.L.; Sotirchos, I.M.; Davey, M.W. The activity and hydrogen peroxide sensitivity of the peroxiredoxins from the parasitic nematode Haemonchus contortus. Mol. Biochem. Parasitol. 2011, 176, 17–24. [Google Scholar] [CrossRef]
- Balasubramanian, N.; Nascimento, G.; Ferreira, R.; Martinez, M.; Simões, N. Pepsin-like aspartic protease (Sc-ASP155) cloning, molecular characterization and gene expression analysis in developmental stages of nematode Steinernema carpocapsae. Gene 2012, 500, 164–171. [Google Scholar] [CrossRef]
- Lin, S.; Jian, H.; Zhao, H.; Yang, D.; Liu, Q. Cloning and characterization of a venom allergen-like protein gene cluster from the pinewood nematode Bursaphelenchus xylophilus. Exp. Parasitol. 2011, 127, 440–447. [Google Scholar] [CrossRef]
- Vandekerckhove, T.T.; Coomans, A.; Cornelis, K.; Baert, P.; Gillis, M. Use of the Verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of “Candidatus xiphinematobacter” cells in nematode hosts. Appl. Environ. Microbiol. 2002, 68, 3121–3125. [Google Scholar] [CrossRef]
- Netto, L.E.; Chae, H.Z.; Kang, S.-W.; Rhee, S.G.; Stadtman, E.R. Removal of hydrogen peroxide by thiol-specific antioxidant enzyme (TSA) is involved with its antioxidant properties TSA possesses thiol peroxidase activity. J. Biol. Chem. 1996, 271, 15315–15321. [Google Scholar] [CrossRef]
- Bruchhaus, I.; Richter, S.; Tannich, E. Removal of hydrogen peroxide by the 29 kDa protein of Entamoeba histolytica. Biochem. J. 1997, 326, 785–789. [Google Scholar]
- Manevich, Y.; Sweitzer, T.; Pak, J.H.; Feinstein, S.I.; Muzykantov, V.; Fisher, A.B. 1-Cys peroxiredoxin overexpression protects cells against phospholipid peroxidation-mediated membrane damage. Proc. Natl. Acad. Sci. 2002, 99, 11599–11604. [Google Scholar]
- Kim, H.-S.; Pak, J.H.; Gonzales, L.W.; Feinstein, S.I.; Fisher, A.B. Regulation of 1-Cys peroxiredoxin expression in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 2002, 27, 227–233. [Google Scholar] [CrossRef]
- Kim, I.-S.; Kim, Y.-S.; Yoon, H.-S. Expression of salt-induced 2-Cys peroxiredoxin from Oryza sativa increases stress tolerance and fermentation capacity in genetically engineered yeast Saccharomyces cerevisiae. Appl. Microb. Biotechnol. 2013, 97, 3519–3533. [Google Scholar] [CrossRef]
- Turturice, B.A.; Lamm, M.A.; Tasch, J.J.; Zalewski, A.; Kooistra, R.; Schroeter, E.H.; Sharma, S.; Kawazu, S.-I.; Kanzok, S.M. Expression of cytosolic peroxiredoxins in Plasmodium berghei Ookinetes is regulated by environmental factors in the mosquito bloodmeal. PLoS Pathog. 2013, 9, e1003136. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Wang, D.; Li, F.; Chi, Y.; Xiang, J. Potential relationship among three antioxidant enzymes in eliminating hydrogen peroxide in penaeid shrimp. Cell Stress Chaperones 2012, 17, 423–433. [Google Scholar] [CrossRef]
- Niu, B.L.; Shen, W.F.; Liu, Y.; Weng, H.B.; He, L.H.; Mu, J.J.; Wu, Z.L.; Jiang, P.; Tao, Y.Z.; Meng, Z.Q. Cloning and RNAi-mediated functional characterization of MaLac2 of the pine sawyer, Monochamus alternatus. Insect Mol. Biol. 2008, 17, 303–312. [Google Scholar] [CrossRef]
- Geldhof, P.; Murray, L.; Couthier, A.; Gilleard, J.S.; McLauchlan, G.; Knox, D.P.; Britton, C. Testing the efficacy of RNA interference in Haemonchus contortus. Int. J. Parasitol. 2006, 36, 801–810. [Google Scholar] [CrossRef]
- Ma, H.; Lu, Q.; Liang, J.; Zhang, X. Functional analysis of the cellulose gene of the pine wood nematode, Bursaphelenchus xylophilus, using RNA interference. Genet. Mol. Res. 2011, 10, 1931–1941. [Google Scholar] [CrossRef]
- Kang, J.S.; Koh, Y.H.; Moon, Y.S.; Lee, S.H. Molecular properties of a venom allergen-like protein suggest a parasitic function in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 2012, 42, 63–70. [Google Scholar] [CrossRef]
- Cheng, X.-Y.; Dai, S.-M.; Xiao, L.; Xie, B.-Y. Influence of cellulase gene knockdown by dsRNA interference on the development and reproduction of the pine wood nematode, Bursaphelenchus xylophilus. Nematology 2010, 12, 225–233. [Google Scholar] [CrossRef]
- Chhabra, A. Advances in genome engineering approaches. Adv. Genet. Eng. 2014, 3, e107. [Google Scholar]
- Buehler, E.; Khan, A.A.; Marine, S.; Rajaram, M.; Bahl, A.; Burchard, J.; Ferrer, M. siRNA off-target effects in genome-wide screens identify signaling pathway members. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef]
- Marine, S.; Bahl, A.; Ferrer, M.; Buehler, E. Common seed analysis to identify off-target effects in sirna screens. J. Biomol. Screen. 2012, 17, 370–378. [Google Scholar] [CrossRef]
- Wang, X.; Chang, L.; Wang, G.; Sun, Z.; Ma, H.; Sun, Q.; Li, J. Protein extraction from the earthworm Eisenia fetida for 2-DE. Proteomics 2010, 10, 1095–1099. [Google Scholar]
- Wang, X.; Chen, Y.; Han, Q.B.; Chan, C.Y.; Wang, H.; Liu, Z.; Cheng, C.H.K.; Yew, D.T.; Lin, M.; He, M.L. Proteomic identification of molecular targets of gambogic acid: Role of stathmin in hepatocellular carcinoma. Proteomics 2009, 9, 242–253. [Google Scholar] [CrossRef]
- De Boer, J.; Yan, Y.; Smant, G.; Davis, E.; Baum, T. In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 1998, 30, 309. [Google Scholar]
- Wang, F.; Wang, Z.; Li, D.; Chen, Q. Identification and characterization of a Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) thermotolerance-related gene: Bx-hsp90. Int. J. Mol. Sci. 2012, 13, 8819–8833. [Google Scholar] [CrossRef]
- Kikuchi, T.; Shibuya, H.; Aikawa, T.; Jones, J.T. Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant-Microbe Interact. 2006, 19, 280–287. [Google Scholar] [CrossRef]
- Urwin, P.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant-Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef]
- Zhu, L.-H.; Ye, J.; Negi, S.; Xu, X.-L.; Wang, Z.-L.; Ji, J.-Y. Pathogenicity of aseptic Bursaphelenchus xylophilus. PLoS One 2012, 7, e38095. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Fu, H.-Y.; Ren, J.-H.; Huang, L.; Li, H.; Ye, J.-R. Screening and Functional Analysis of the Peroxiredoxin Specifically Expressed in Bursaphelenchus xylophilus—The Causative Agent of Pine Wilt Disease. Int. J. Mol. Sci. 2014, 15, 10215-10232. https://doi.org/10.3390/ijms150610215
Fu H-Y, Ren J-H, Huang L, Li H, Ye J-R. Screening and Functional Analysis of the Peroxiredoxin Specifically Expressed in Bursaphelenchus xylophilus—The Causative Agent of Pine Wilt Disease. International Journal of Molecular Sciences. 2014; 15(6):10215-10232. https://doi.org/10.3390/ijms150610215
Chicago/Turabian StyleFu, Han-Yu, Jia-Hong Ren, Lin Huang, Hao Li, and Jian-Ren Ye. 2014. "Screening and Functional Analysis of the Peroxiredoxin Specifically Expressed in Bursaphelenchus xylophilus—The Causative Agent of Pine Wilt Disease" International Journal of Molecular Sciences 15, no. 6: 10215-10232. https://doi.org/10.3390/ijms150610215