1. Introduction
Epithelial ovarian carcinomas (EOC) are composed of a group of heterogeneous subtypes classified by their histology and the degree of epithelial proliferation and invasion. Clear cell (CCC), endometrioid (EC), mucinous (MC) and high-grade serous carcinoma (SC) are four common subtypes of EOC. Within the four subtypes, high-grade SC is the most common type accounting for 70% of EOC, followed by CCC, while MC is relatively rare. However, the carcinogenesis of EOC is still poorly understood. Based on the clinicopathological and molecular features, the dualistic model was proposed and divided EOCs into type I and II categories [
1]. The type I EOC, including CCC, EC and MC, usually originating from the mutations of KRAS, BRAF, ERBB2, CTNNB1, PTEN and PIK3CA, is genetically stable and has a relatively indolent behavior [
2]. The type II EOC, mainly high-grade SC, displays TP53 mutation in over 80% of the cases, exhibits impaired DNA damage repair and has a more uncontrolled cell differentiation and aggressive behavior. This hypothesis was based on the studies performed in the author’s laboratory and correlated with the clinical, pathologic and molecular features of the disease. However, there is no single study, nor integrative analysis to demonstrate this hypothesis and compare the pathogenesis among the four EOC subtypes. As a result, we conducted a gene set-based analysis integrating the microarray gene expression profiles of the four EOC subtypes from the publicly available database. Gene expression microarray is the primary tool for investigating cancers, the analysis of gene expression profiles usually starts with detecting the differentially expressed genes (DEG) by statistical methods, and then the aberrant Gene Ontology (GO) terms or signaling pathways are inferred from the DEGs. This workflow identifies the most significant disease-related genes, function or processes annotated by GO terms or signaling pathways, however, it will focus only on the significant ones and omit those whose
p values do not reach statistical significance. In fact, genes or GO terms that did not reach the significance also play a role in the carcinogenesis of EOCs. Besides, only limited functions defined by the GO term or canonical pathways are analyzed; the complete information about the regulation of the functions i.e., functionome in EOC is not provided. To address these limitations, we investigated the pathogenesis of the four subtypes of EOC with microarray gene expression profiles of EOC and their functionomes. The biological function was quantized by converting the gene expression profiles to a gene set regularity (GSR) index computed by modifying the DIRAC algorithm [
3], which measured the matching degree of gene expression rankings in a given gene set between two different phenotypes, i.e., EOC and the normal ovarian tissue control in this study. This model utilized the gene set definitions from the GO term [
4] and Reactome pathway [
5] databases downloaded from the Molecular Signatures Database (MSigDB) [
6]. These two gene set definitions collect relatively comprehensive biological functions, processes or signaling pathways. We then utilized them to annotate human functionomes. The GO database contains 1454 gene sets, defining biological functions, process and cellular components; the canonical pathway database contains 1330 curated canonical signaling pathways. In our previous study [
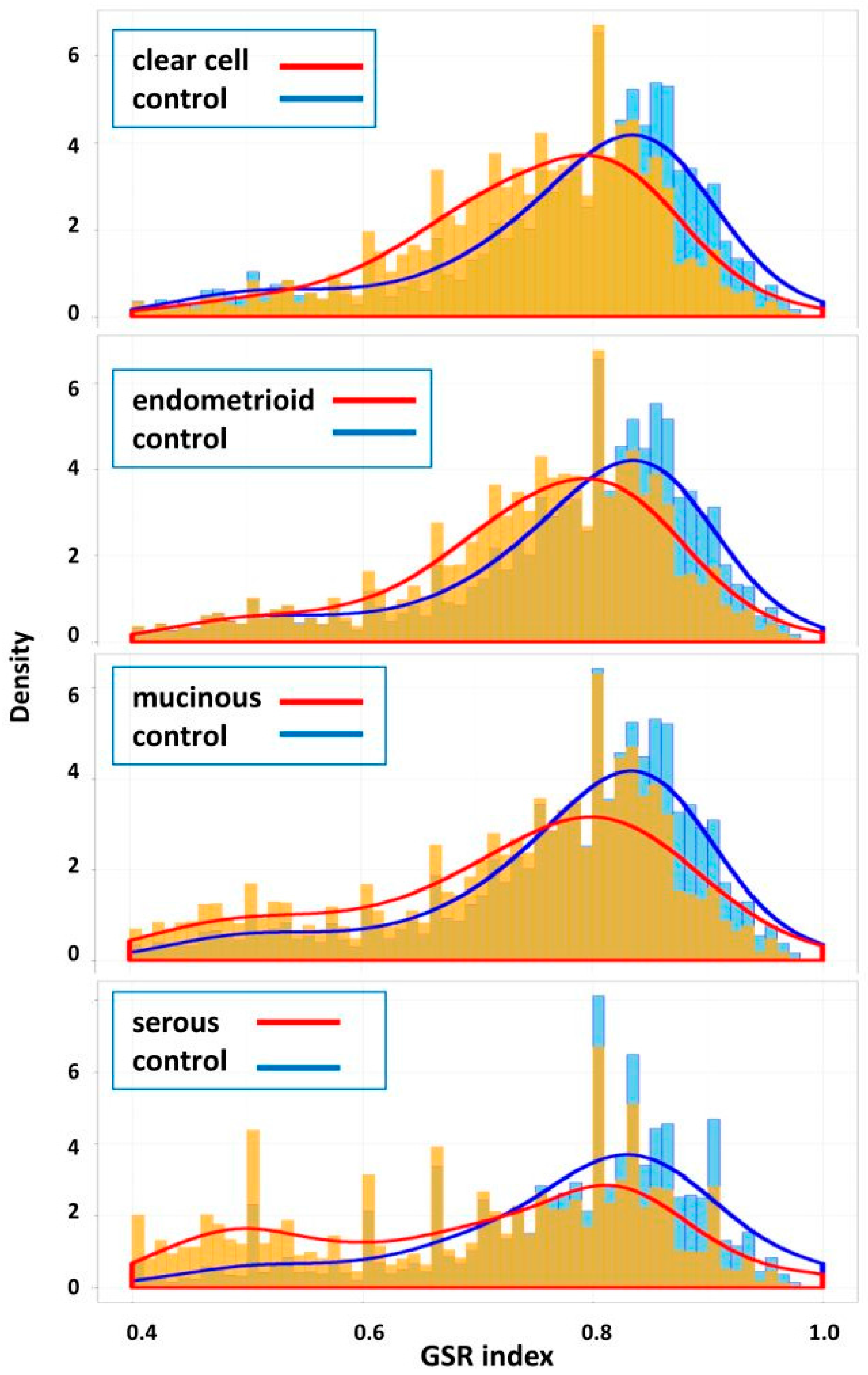
7], we demonstrated by the GSR indices a stepwise deterioration of cellular function regularity during SC progression from stage I to stage IV according to International Federation of Gynecology and Obstetrics (FIGO). The pathogenesis of SC centered on cell cycle deregulation accompanied with multi-functional aberrations and interactions. To further explore the pathogenesis and relationship among different subtypes of EOCs, we collected the gene expression datasets of the four common subtypes of EOC and normal ovarian samples from the publicly available databases and converted them into the GSR indices, ranging from 0 to 1 and reflecting the regularities of functions defined by the GO terms or Reactome pathways. Then, the pathogenesis of the four EOC subtypes was investigated and compared with the GSR indices by hierarchical clustering, statistical methods and exploratory factor analysis (EFA).
3. Discussion
Cancers are usually involved in multiple aberrations of gene and function as well as their interactions. In order to take these features into consideration, we utilized the GSR model to investigate the function regularities in cancers. Instead of detecting the DEGs, the model starts with converting the microarray gene expression profiles into quantized biological functions through a list of gene sets defined by the GO terms or Reactome pathways, and then the pathogenesis is evaluated by comparing the differences of functional regulation between the cases with the normal control groups. These quantized regularities of functions, i.e., the GSR indices, are computed by the modified DIRAC algorithm, which converts the gene expression levels to a gene expression ranking list in a gene set, and then measures the matching degree of gene expression rankings between two different phenotypes. We utilized a baseline gene set expression ranking template, defined as the most common gene expression ranking in the normal control populations for each gene set, as a standard to measure the regularity of gene ranking in either EOC or normal ovarian control sample. Then, the GSR index is computed by measuring the matching degree between the gene expression rankings of each ovarian cancer or normal ovarian control sample with the baseline gene set expression ranking template for each gene set. After being standardized by the baseline gene set template, the GSR indices of the four EOC subtypes can be compared based on the same standard. Besides, the GSR indices are computed based on the gene expression rankings; the gene expression levels are converted into ordinal data, and the ordinal data will encounter less cross-platform bias than the gene expression levels during integrating the datasets from different DNA microarray platforms. Computing the gene expression ranking in a gene set will take the gene interactions in a gene set into consideration. In contrast to the “genome” analyzed with gene expression microarray, this model investigates “functionome” with the GSR indices. By converting tens of thousands of gene expression profiles to approximately one thousand GSR indices, this approach will diminish the data noise, simplify the complexity of the subsequent analyses, and facilitate the performance of machine leaning. Besides, each GSR index is normalized to a value ranging from 0 to 1, in favor of the subsequent analyses.
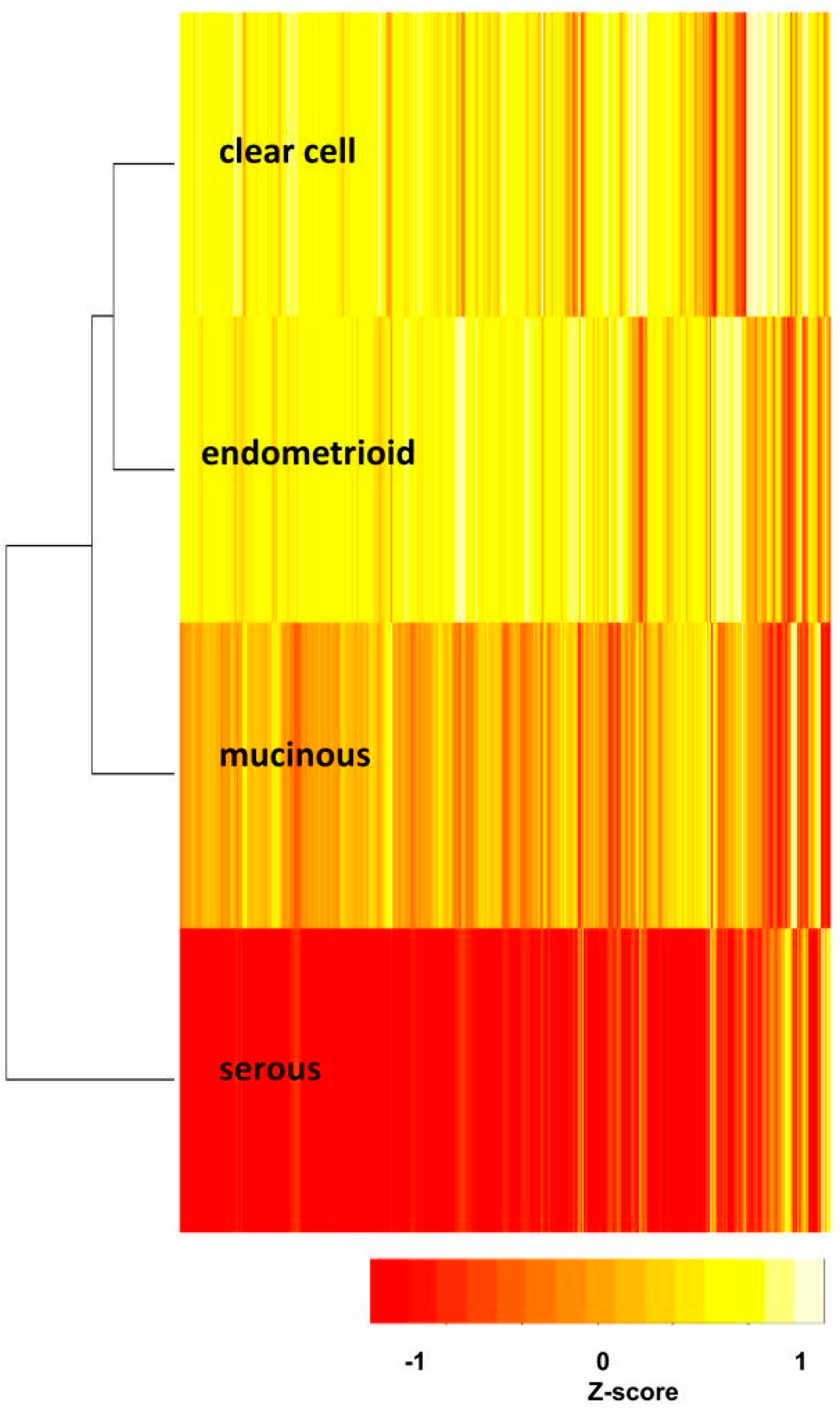
The functionome of each subtype was computed through either GO term or Reactome pathway gene set database, both databases collect relative comprehensive human biological functions and processes, and provide the browsers for viewing the hierarchy of GO terms (AmiGO 2) [
9] and pathways (Reactome Pathway Browser) [
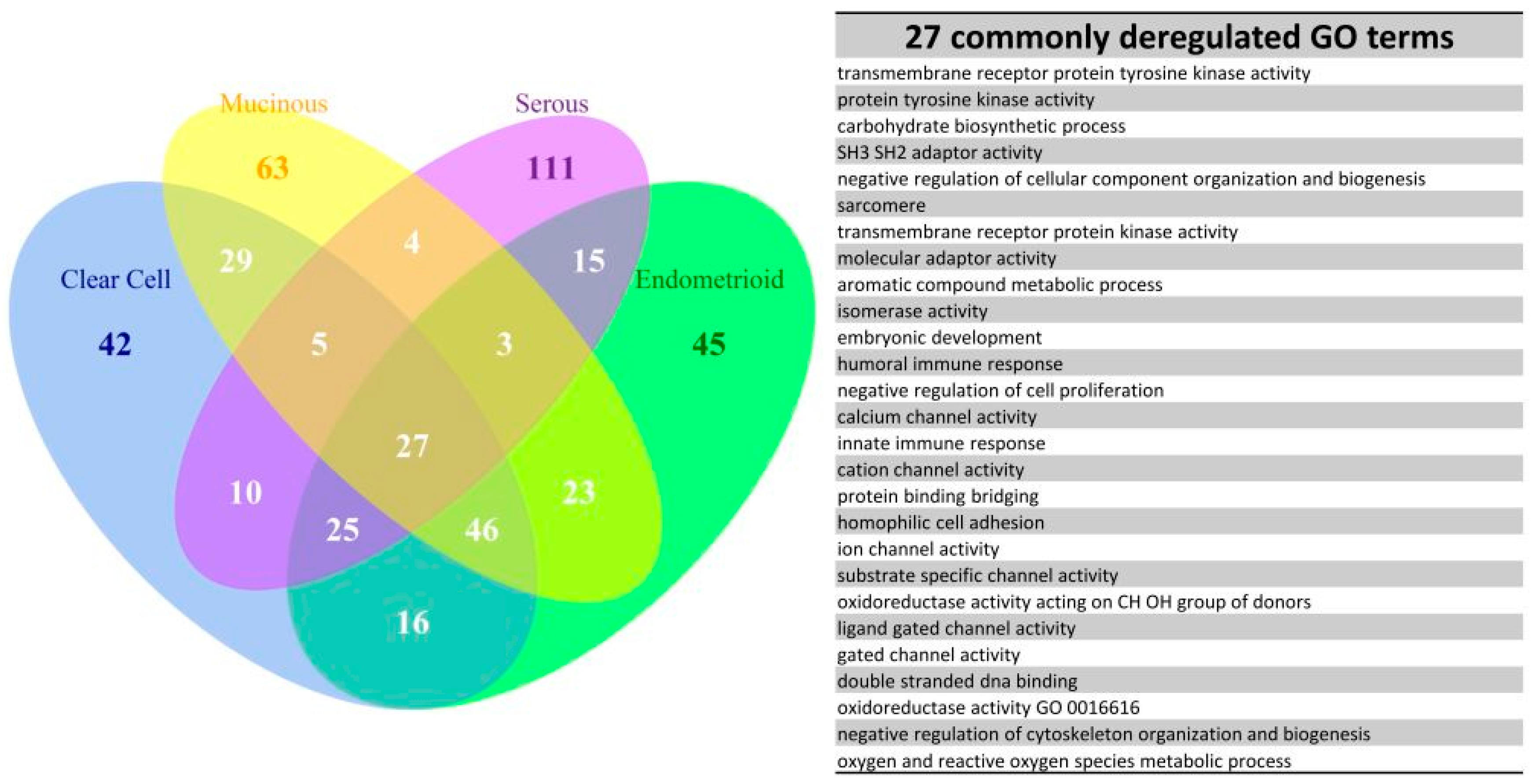
10], facilitating the clarification of the relationships among numerous deregulated GO terms or pathways. The functionome was composed of approximately 1400 GO or 600 Reactome GSR indices for each case, when displayed on the heatmap, the functionomes of the four EOC subtypes could be visualized and show distinguishable patterns. These patterns could be recognized, classified and predicted by the machine learning. Our result revealed excellent binary or multiclass classification; it implied that the functionomes composed of GSR indices could be utilized as the basis of molecular classification by machine learning. Subsequently, the pathogenesis of the four subtypes was investigated by evaluating the GSR indexes. From the results of histograms and hierarchical clustering among the four subtypes, it could be found that CCC and EC had the closest relationship, followed by MC, and SC was relatively different from the others in terms of functional regulations. Indeed, the four subtypes shared quite a number of common deregulated functions, including cell adhesion, oxidoreductase activity, protein binding, channel activity and metabolism. However, deregulations of chromatin assembly, ERBB, PI3K-AKT pathways were more common among CCC, EC and MC but not in SC. In contrast, the predominant deregulated functions in SC were cell cycle control.
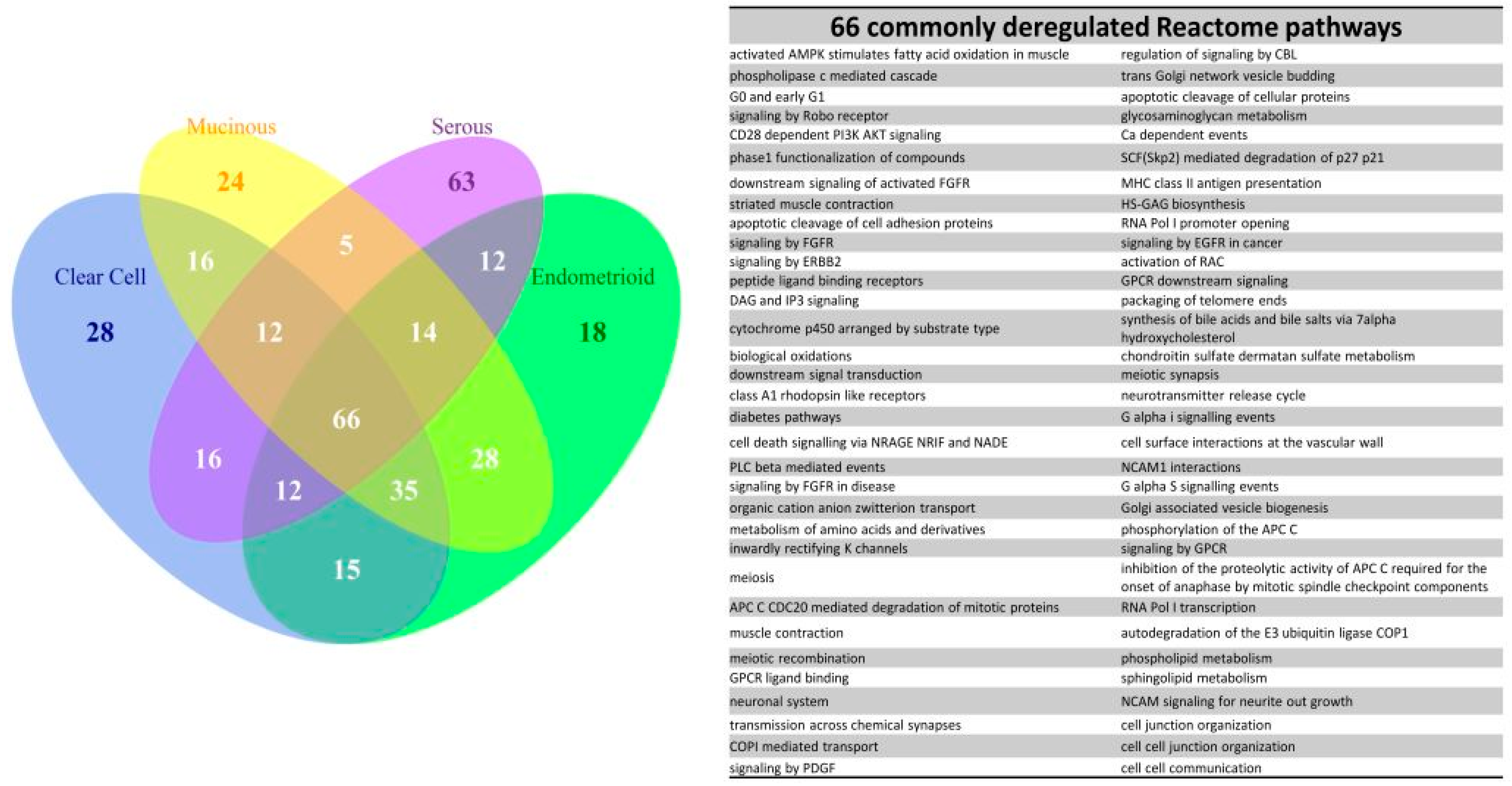
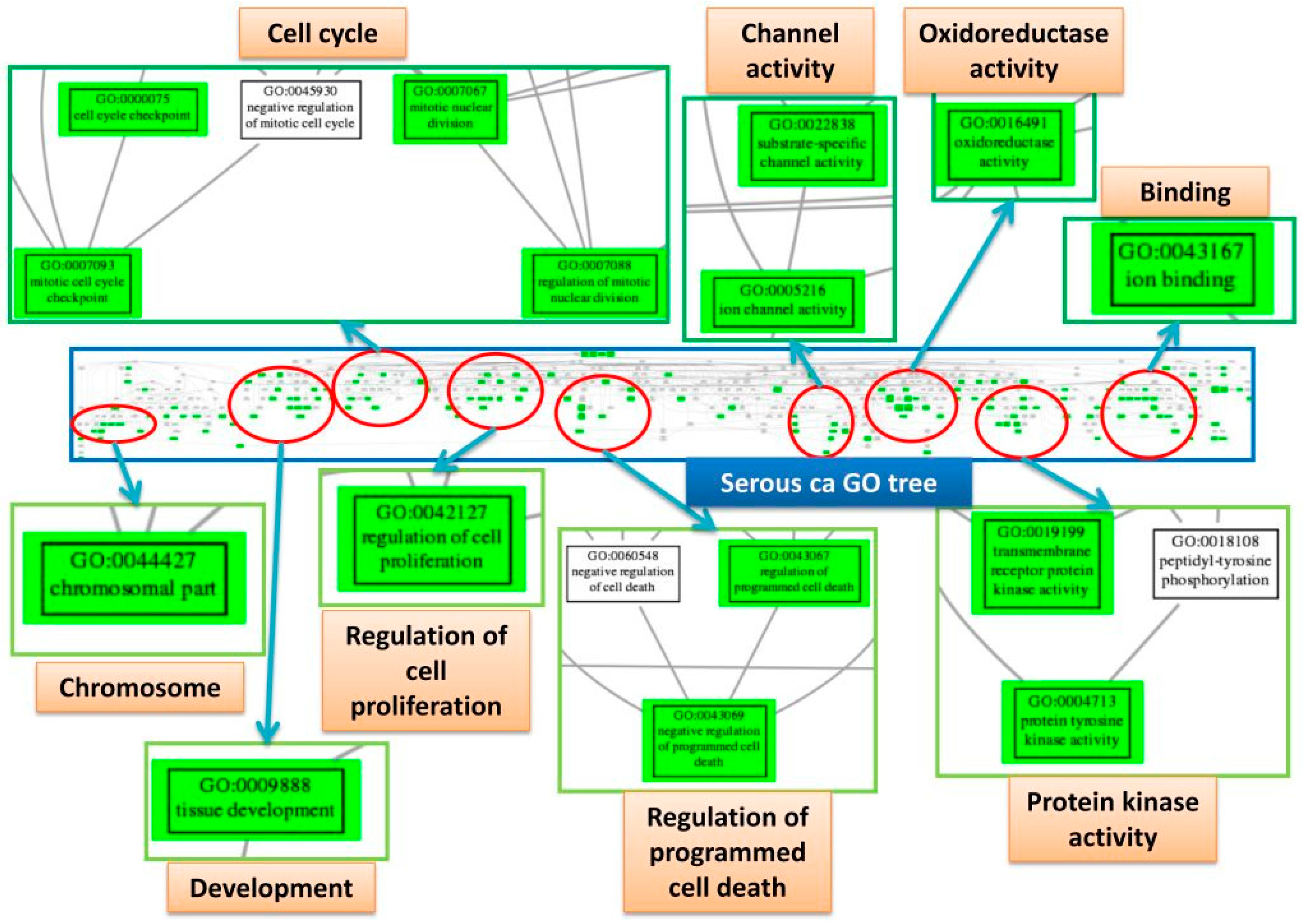
We further explored the pathogenesis and the relationship among the four subtypes by the EFA. The results of EFA using GO terms disclosed that CCC, EC and MC shared a similar structure of pathogenesis, associated with binding, channel activity, cell adhesion, oxidoreductase activity, protein kinase activity, G protein activity and chromatin assembly. The results of EFA using Reactome pathway gene sets revealed the common deregulation of the PI3K-AKT and ERBB pathways. In contrast, the results of EFA for the SC group revealed the pathogenesis mainly involved in apoptosis, mitosis and cell division and cell cycle checkpoint. Overlapped deregulated functions among the four EOC subtype groups were also found, such as protein tyrosine kinase activity, carbohydrate biosynthetic process, immune response, channel activity, cell adhesion and oxidoreductase activity. The channel activity was demonstrated to be involved in the cell cycle control in the carcinogenesis of EOC [
11], and cell adhesion played an important role in the metastasis of EOC [
12]. These findings draw the conclusion that the two overlapped, but distinguishable function regulation patterns existing among the four subtypes of EOC. The first pattern observed in the CCC, EC and MC groups had moderate, deregulated functions involved in oxidoreductase activity, channel activity, binding activity, metabolism, chromatin assembly, cell adhesion, PI3K-AKT and ERBB signaling pathway. The secondary pattern, observed in the SC groups, had more severe functional regularity and was predominantly involved in the cell cycle deregulation. These two function regulation patterns were compatible with the type I and type II classifications proposed by the dualistic model of ovarian carcinogenesis: the type I EOCs, including CCC, EC and MC, usually originated from the mutation of KRAS, BRAF, ERBB2, PTEN and PIK3CA, are genetically stable and have a relatively indolent behavior; the type II EOCs, mainly high-grade SC, primarily exhibit a TP53 signature, have a more uncontrolled cell cycle and aggressive behavior. The type I and II EOCs were compatible with the first and second patterns of function regulation in our study, respectively.
This study also showed evidence disclosing the relationship between deregulated functions and carcinogenesis. The association of CCC and EC with endometriosis has been repeated reported [
13,
14]. The cells in the endometriosis foci will be exposed to the reactive oxygen species (ROS) and are subjected to more DNA damage [
15]. As the dendrogram showed in this study, the CCC and EC groups exhibited a relatively close relationship and shared many commonly deregulated GO terms, such as oxidoreductase activity and cell adhesion; both are the characteristic features of the pathogenesis of endometriosis. These findings provided the evidence supporting the role of endometriosis during the carcinogenesis of CCC and EC.
Our results showed the PI3K-AKT signaling pathway was a key element of the pathogenesis of EOCs. PI3K-AKT has been demonstrated to play an important role in the carcinogenesis of EOC, especially in CCC and EC. The deregulation of this signaling pathway may be originated from the loss of PTEN in 40% cases [
16], PIK3CA mutation in 33% cases [
17] or AKT amplification in 14% cases [
18] of CCC patients. PI3K is the major downstream effector of receptor tyrosine kinases (RTK) and GPCR. If PI3K is activated, apoptosis will be inhibited and leads to cell proliferation [
19]. Both of PI3K-AKT and G protein deregulation were detected with statistical significances in this study. As the results of CCC-EC-MC combined analysis listed in the
Table S9, the GO terms “inositol or phosphatidylinositol phosphatase activity” and “transmembrane receptor protein tyrosine kinase activity” were the first and sixth top deregulated GO gene sets. ERBB2 was the first deregulated pathways for CCC and EC, its expression in EOC varies widely, ranging from 20% to 30% of cases [
20]. ERBB is a member of the epidermal growth factor receptor (EGFR) family, it can activate the PI3K-AKT pathway and may represent a prognostic factor in primary EOC [
21]. The 9th deregulated Reactome pathway “PI3K events in ERBB2 signaling” in the CCC-EC-MC combined group indicated the interaction between the two important deregulated Reactome pathways in the carcinogenesis of EOC (
Table S10).
However, there are limitations when applying the GSR model to investigate the carcinogenesis of EOCs. As an illustration, the TP53 mutation is a common aberration in high-grade SC. The gene set related to TP53 could be found in the list of Reactome pathway database; however, they did not appear on the top of the significantly deregulated pathway list in this study; the first one appearing on the list was the 122th gene set “P53 dependent G1 DNA damage response” with a p value of 4.02 × 10−17. This finding illustrates the first limitation of this model: if the level of gene expression change does not reach the required extent, the gene expression ranking as well as the GSR index will remain unchanged and the aberration could not be detected. The second limitation is the incompleteness of gene set definitions. For example, there was no definition of PTEN gene set in the GO and Reactome gene set database, so no PTEN aberration was found in this study, although this model discovered a lot of PI3K related functions and pathway aberrations because the PI3K were the effector of PTEN. The third limitation is the false positivity. The third most deregulated Reactome pathway in the MC group was “olfactory signaling pathway” with a p value of 1.32 × 10−12, which should be independent of the carcinogenesis of MC. This situation can be checked and clarified via the Reactome Pathway Browser. When mapping to the browser, the hierarchy showed the “olfactory signaling pathway” was a member of the GPCR signaling pathway and contained elements involved with the regulation of G protein, and G protein was shown to play an important role in the carcinogenesis of EOC in this study. This false positivity happened because of the presence of the G protein-related gene elements in the gene set. Another limitation of this study was that the DEGs derived from the integrative analysis had not been validated. One of the best ways to validate these DEGs is RNA seq or protein expression for the samples of the four EOC subtypes. We attempted to validate the DEGs in our study by collecting the RNA seq datasets for the four EOC subtypes from two important publically available databases: The Cancer Genome Atlas (TCGA) and NCBI Sequence Read Archive (SRA). However, this validation was not feasible because the available samples of CCC, EC and MC were not enough to get significant statistical significance. Further investigation is still needed for validation of these DEGs.