Preparation of A Spaceflight: Apoptosis Search in Sutured Wound Healing Models
Abstract
:1. Introduction
2. Results and Discussion
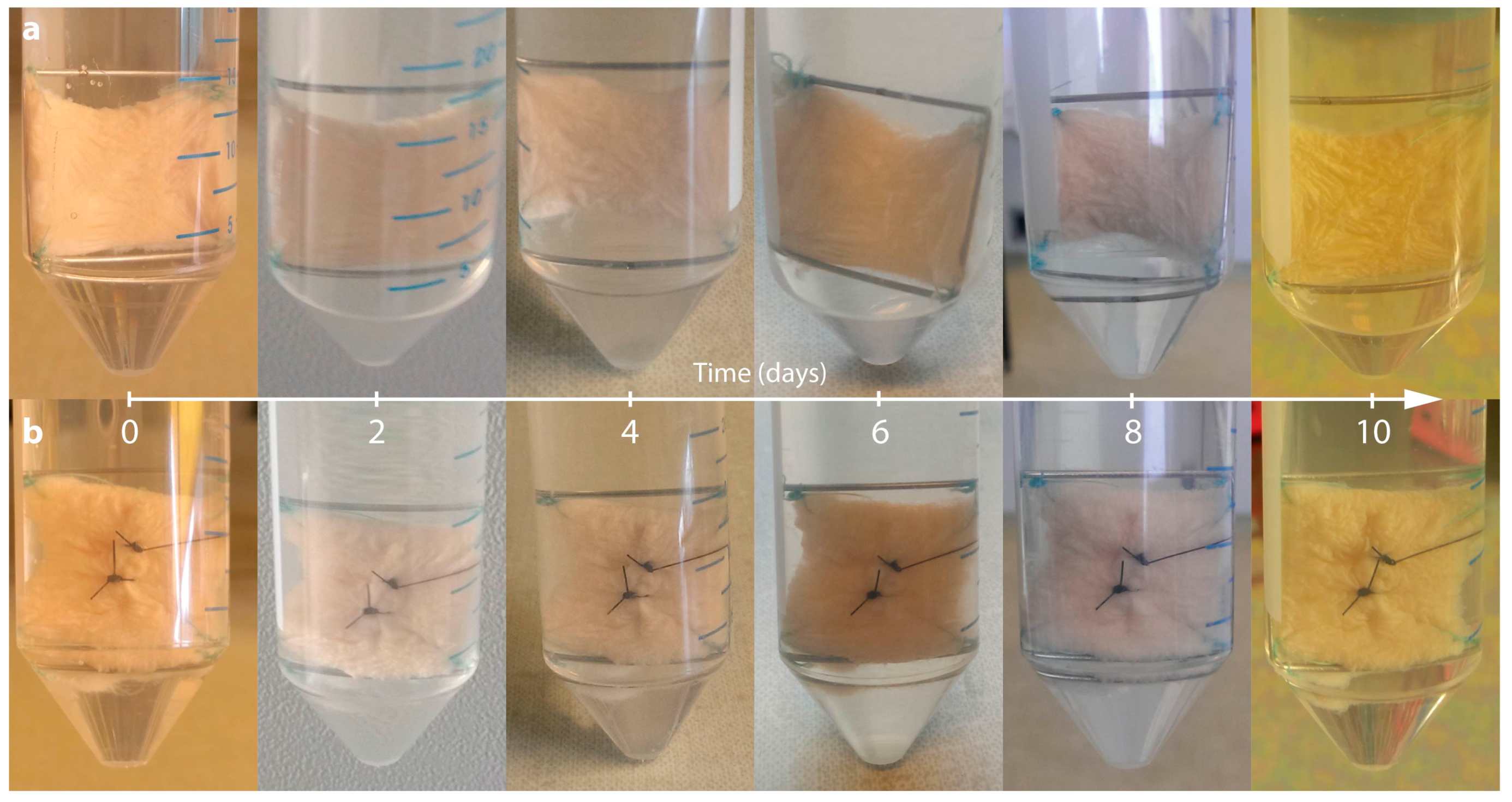
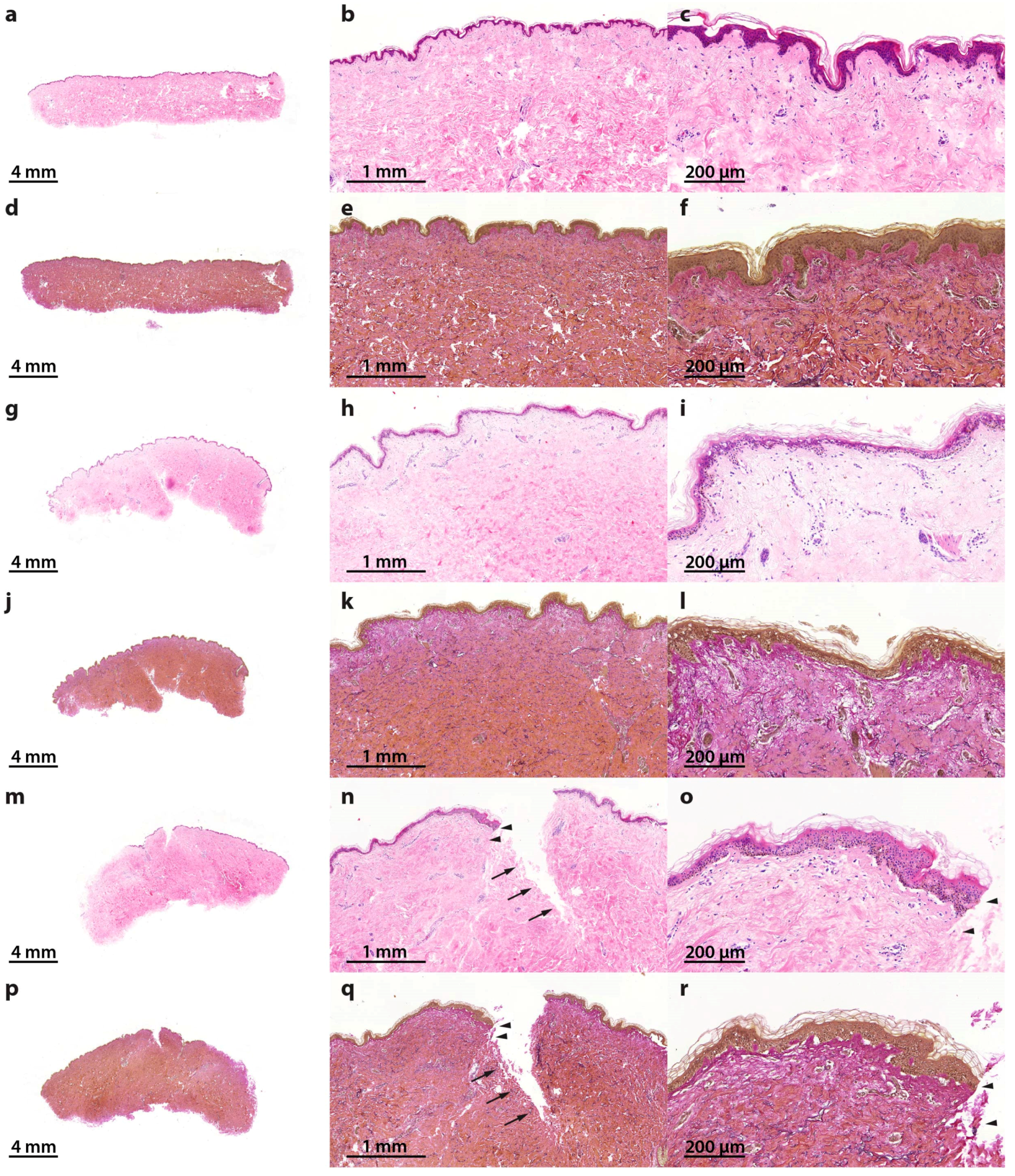
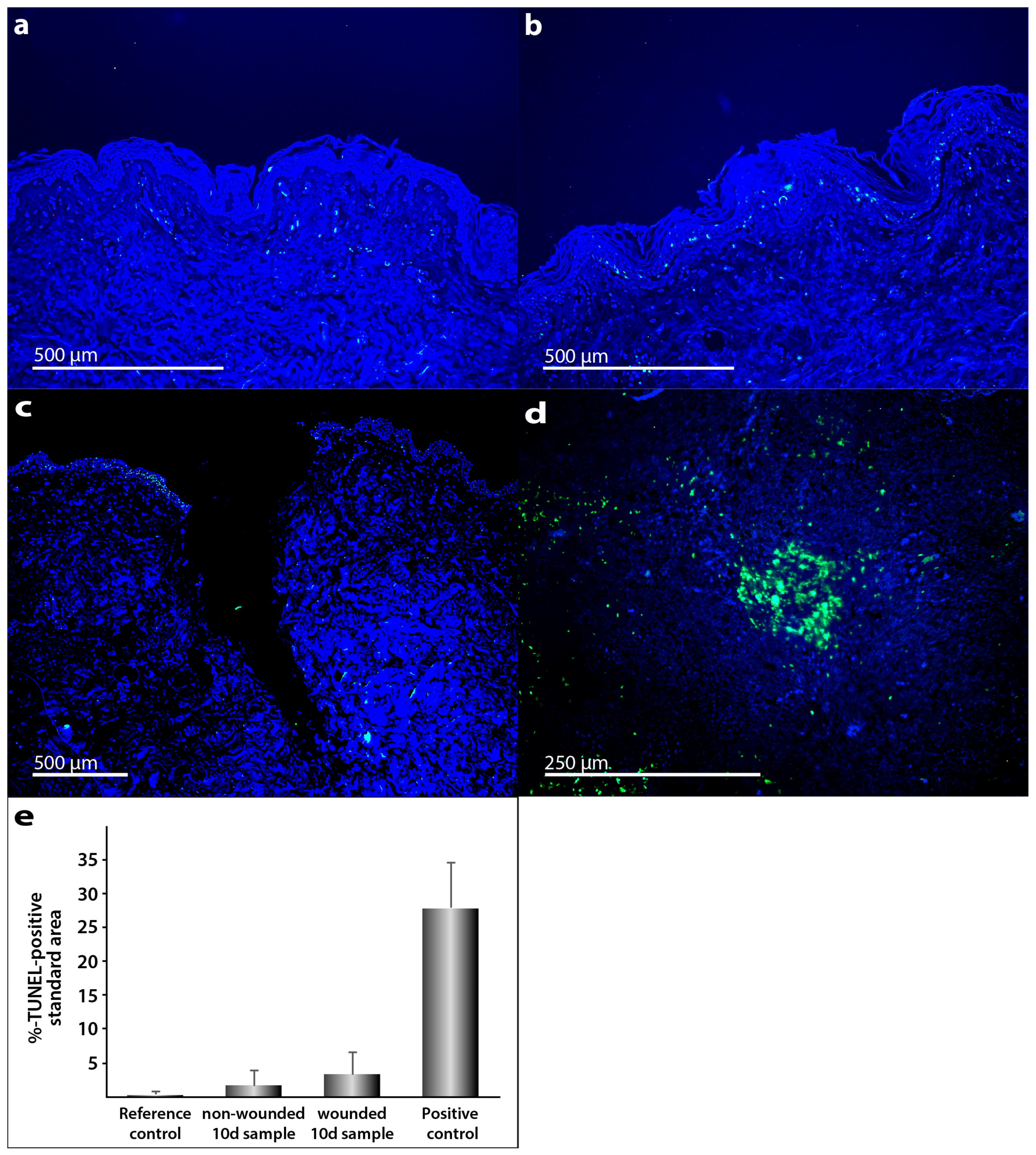
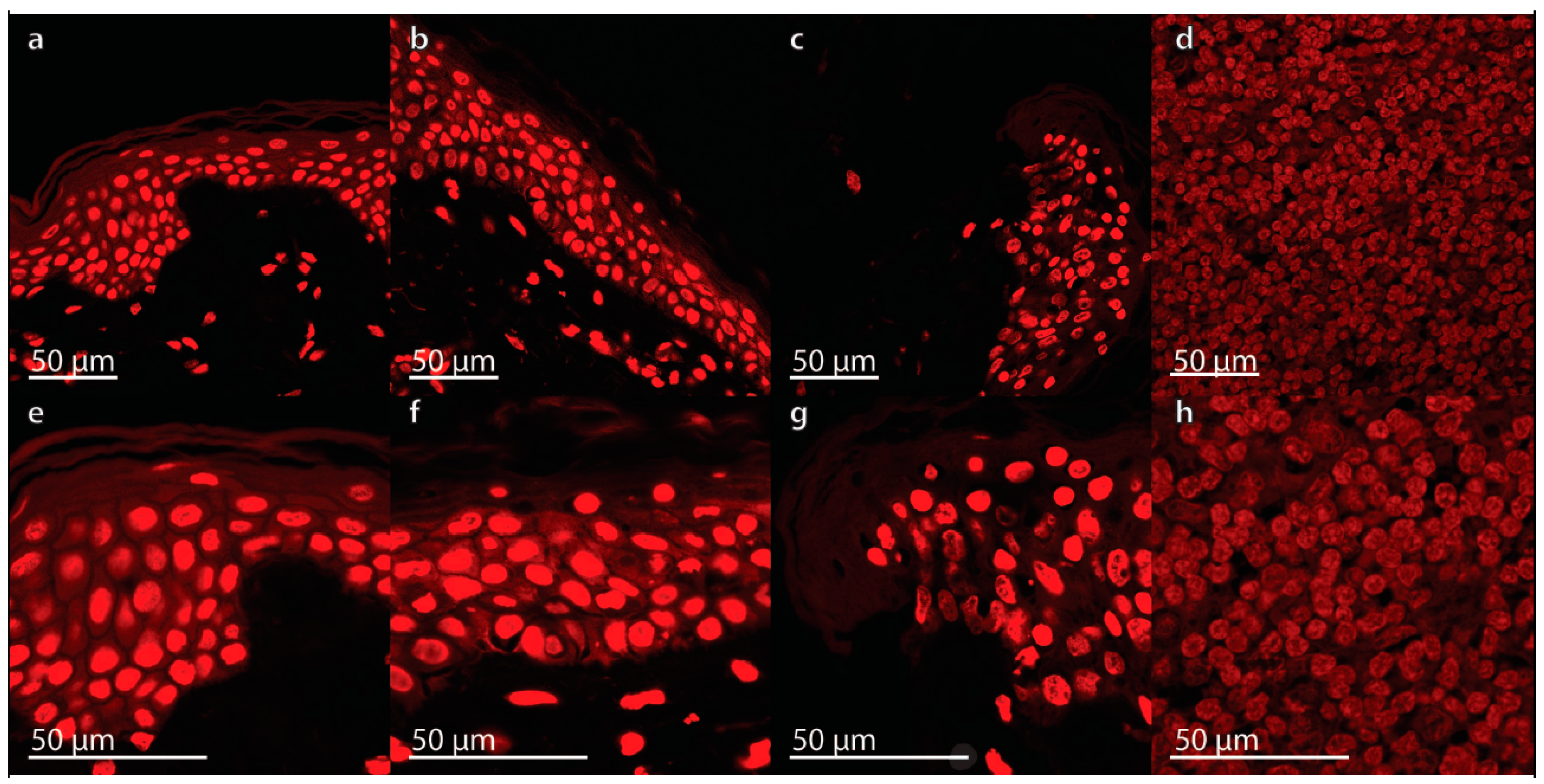
2.1. Characterization of the Morphological Changes and Testing the Viability of the Tissue Specimens
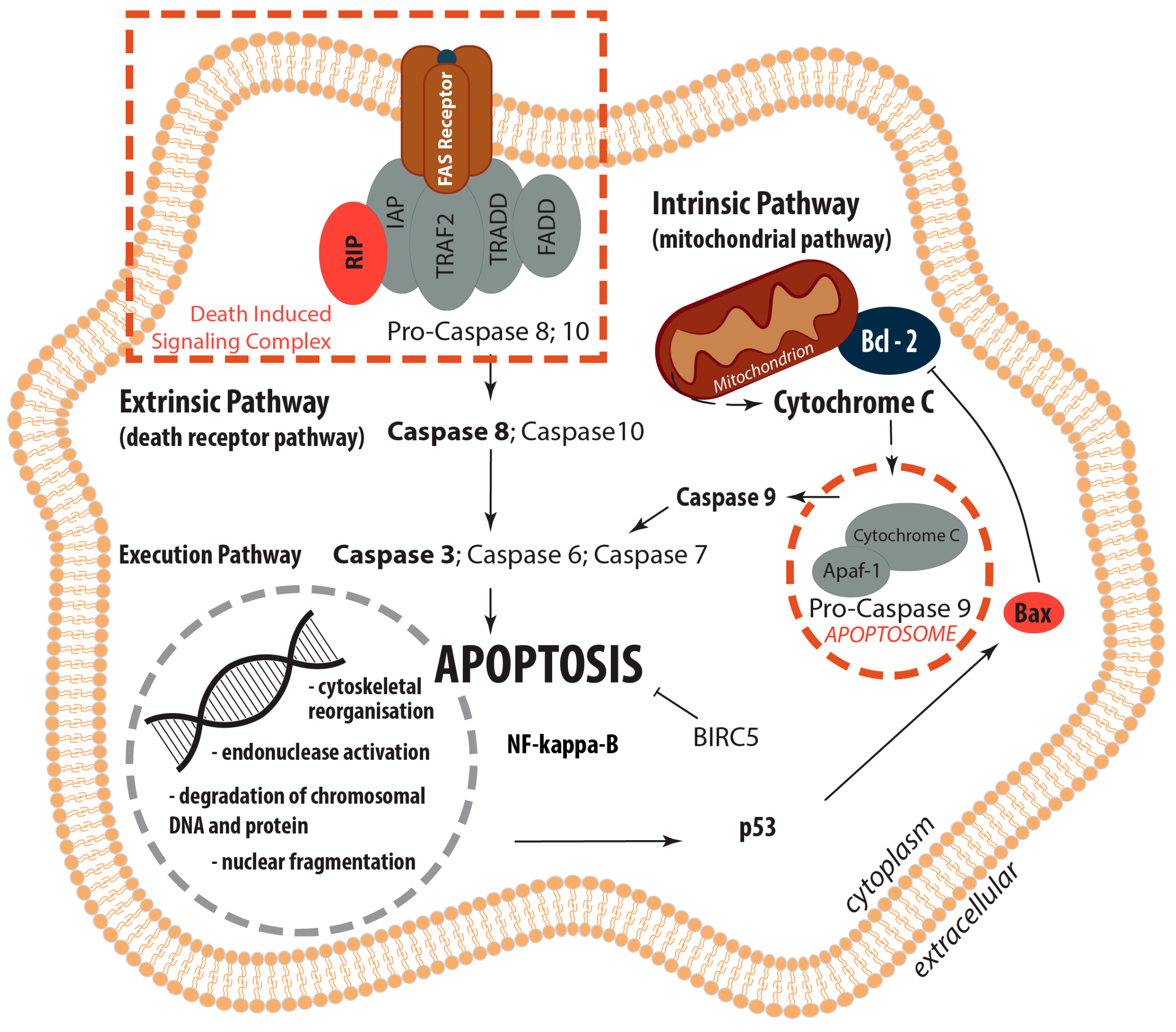
2.2. Determination of the Gene Expression of Activators and Inhibitors of Apoptosis (Extrinsic and Intrinsic Pathways of Apoptosis) and Extracellular Matrix Components in Wound and Suture Models, Including Analysis of the Interaction Network of the Genes of Interest
2.2.1. Gene Expression of Activators and Inhibitors of Apoptosis
2.2.2. Gene Expression Profile of Extracellular Matrix Proteins
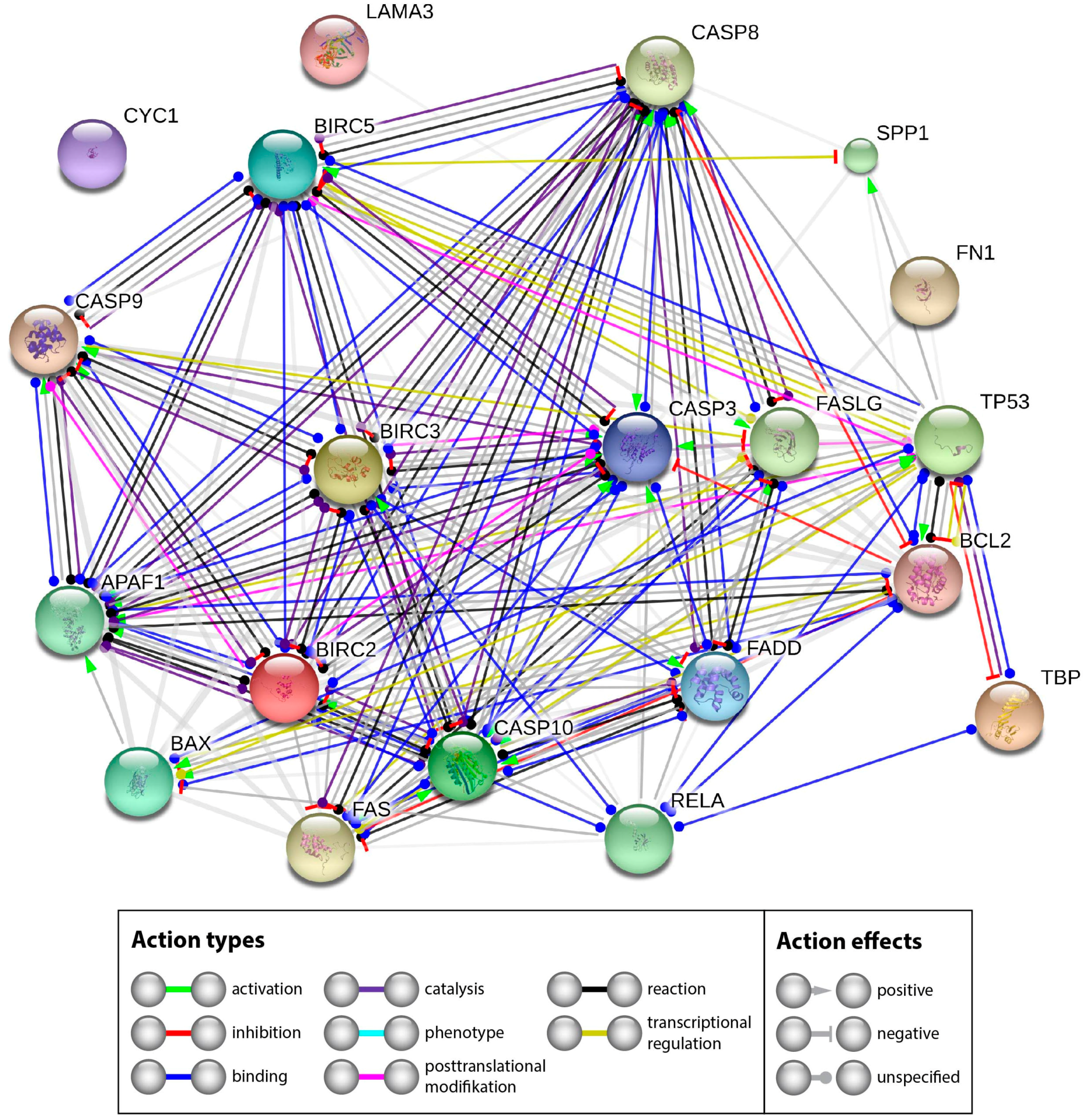
2.2.3. Interaction Network of the Genes of Interest
2.3. Design of the Analytical Protocols to Be Performed in the Future on Tissue Specimens after the Post-Spaceflight Download of the Samples
3. Materials and Methods
3.1. Media, Supplements and Reagents
3.2. Tissue Preparation and Organ Culture
3.3. RNA Extraction and qPCR
3.4. Histochemistry and Microscopy
3.5. TUNEL Assay
3.6. Propidium Iodide Staining
3.7. Western Blot Analysis
3.8. STRING Analysis
3.9. Statistical Evaluation
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| APAF-1 | Apoptotic protease-activating factor 1 |
| BIRC5 | Apoptosis inhibitor surviving |
| BAX | Apoptosis regulator Bax (BCL2 associated athanogene) gene |
| BCL2 | Apoptosis regulator Bcl-2 (B-cell lymphoma protein 2) gene |
| Caspase-3 | Cysteinyl aspartic acid-protease-3 |
| Caspase-6 | Cysteinyl aspartic acid-protease-6 |
| Caspase-7 | Cysteinyl aspartic acid-protease-7 |
| Caspase-8 | Cysteinyl aspartic acid-protease-8 |
| Caspase-9 | Cysteinyl aspartic acid-protease-9 |
| Caspase-10 | Cysteinyl aspartic acid-protease-10 |
| CYC1 | Cytochrome C gene |
| DAPI | 4′,6-Diamidin-2-phenylindol |
| ECM | Extracellular Matrix |
| FADD | Fas-associated death domain |
| FAS/CD95 | Tumor necrosis factor receptor superfamily member 6 |
| FN1 | Fibronectin gene |
| IAP | Inhibitor of Apoptosis Proteins |
| RIP | Cell death protein RIP |
| TNFA | Tumor necrosis factor α |
| TNFB | Lymphotoxin-α |
| TRAF2 | TNF receptor-associated factor 2 |
| TRAIL | Tumor necrosis factor ligand superfamily member 10 |
| TUNEL | Terminal deoxynucleotidyl transferase dUTP nick end labeling |
References
- Singer, A.J.; Clark, R.A. Cutaneous wound healing. N. Engl. J. Med. 1999, 341, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Eming, S.A.; Martin, P.; Tomic-Canic, M. Wound repair and regeneration: Mechanisms, signaling, and translation. Sci. Transl. Med. 2014, 6, 265sr6. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Reinke, J.M.; Sorg, H. Wound repair and regeneration. Eur. Surg. Res. 2012, 49, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Darby, I.A.; Laverdet, B.; Bonte, F.; Desmouliere, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014, 7, 301–311. [Google Scholar] [PubMed]
- Greenhalgh, D.G. The role of apoptosis in wound healing. Int. J. Biochem. Cell Biol. 1998, 30, 1019–1030. [Google Scholar] [CrossRef]
- Huang, N.F.; Zac-Varghese, S.; Luke, S. Apoptosis in skin wound healing. Wounds 2003, 15, 182–194. [Google Scholar]
- Drudi, L.; Ball, C.G.; Kirkpatrick, A.W.; Saary, J.; Grenon, S.M. Surgery in space: Where are we at now? Acta Astronaut. 2012, 79, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Davidson, J.M.; Aquino, A.M.; Woodward, S.C.; Wilfinger, W.W. Sustained microgravity reduces intrinsic wound healing and growth factor responses in the rat. FASEB J. 1999, 13, 325–329. [Google Scholar] [PubMed]
- Campbell, M.R.; Williams, D.R.; Buckey, J.C.; Kirkpatrick, A.W. Animal surgery during spaceflight on the Neurolab Shuttle mission. Aviat. Space Environ. Med. 2005, 76, 589–593. [Google Scholar] [PubMed]
- Midura, R.J.; Su, X.; Androjna, C. A simulated weightlessness state diminishes cortical bone healing responses. J. Musculoskelet. Neuronal Interact. 2006, 6, 327–328. [Google Scholar] [PubMed]
- Delp, M.D. Unraveling the complex web of impaired wound healing with mechanical unloading and physical deconditioning. J. Appl. Physiol. 2008, 104, 1262–1263. [Google Scholar] [CrossRef] [PubMed]
- Radek, K.A.; Baer, L.A.; Eckhardt, J.; DiPietro, L.A.; Wade, C.E. Mechanical unloading impairs keratinocyte migration and angiogenesis during cutaneous wound healing. J. Appl. Physiol. 2008, 104, 1295–1303. [Google Scholar] [CrossRef] [PubMed]
- Heinemeier, K.M.; Olesen, J.L.; Haddad, F.; Schjerling, P.; Baldwin, K.M.; Kjaer, M. Effect of unloading followed by reloading on expression of collagen and related growth factors in rat tendon and muscle. J. Appl. Physiol. 2009, 106, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Morbidelli, L.; Monici, M.; Marziliano, N.; Cogoli, A.; Fusi, F.; Waltenberger, J.; Ziche, M. Simulated hypogravity impairs the angiogenic response of endothelium by up-regulating apoptotic signals. Biochem. Biophys. Res. Commun. 2005, 334, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Monici, M.; Cialdai, F.; Romano, G.; Fusi, F.; Egli, M.; Pezzatini, S.; Morbidelli, L. An in Vitro Study on Tissue Repair: Impact of Unloading on Cells Involved in the Remodelling Phase. Microgravity Sci. Technol. 2011, 23, 391–401. [Google Scholar] [CrossRef]
- Cialdai, F.; Vignali, L.; Morbidelli, L.; Colciago, A.; Celotti, F.; Santi, A.; Caselli, A.; Cirri, P.; Monici, M. Modeled Microgravity Affects Fibroblast Functions Related to Wound Healing. Microgravity Sci. Technol. 2017, 29, 121–132. [Google Scholar] [CrossRef]
- Hautier, A.; Sabatier, F.; Stellmann, P.; Andrac, L.; De Gorce, Y.N.; Dignat-George, F.; Magalon, G. Assessment of organ culture for the conservation of human skin allografts. Cell Tissue Bank. 2008, 9, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Gawlitta, D.; Oomens, C.W.; Baaijens, F.P.; Bouten, C.V. Evaluation of a continuous quantification method of apoptosis and necrosis in tissue cultures. Cytotechnology 2004, 46, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Elson, K.M.; Fox, N.; Tipper, J.L.; Kirkham, J.; Hall, R.M.; Fisher, J.; Ingham, E. Non-destructive monitoring of viability in an ex vivo organ culture model of osteochondral tissue. Eur. Cell Mater. 2015, 29, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Lenselink, E.A. Role of fibronectin in normal wound healing. Int. Wound J. 2015, 12, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Rai, N.K.; Tripathi, K.; Sharma, D.; Shukla, V.K. Apoptosis: A basic physiologic process in wound healing. Int. J. Low. Extrem. Wounds 2005, 4, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.S.; Chen, S.N. Apoptotic cell: Linkage of inflammation and wound healing. Front. Pharmacol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kane, C.D.; Greenhalgh, D.G. Expression and localization of p53 and BCL-2 in healing wounds in diabetic and nondiabetic mice. Wound Repair Regen. 2000, 8, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Kleszczynski, K.; Fischer, T.W. Development of a short-term human full-thickness skin organ culture model in vitro under serum-free conditions. Arch. Dermatol. Res. 2012, 304, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Oliver, N.; Babu, M.; Diegelmann, R. Fibronectin gene transcription is enhanced in abnormal wound healing. J. Investig. Dermatol. 1992, 99, 579–586. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Carraher, C.; Schwarzbauer, J.E. Assembly of fibronectin extracellular matrix. Annu. Rev. Cell Dev. Biol. 2010, 26, 397–419. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Jabusch, H.C.; Kossmehl, P.; Huber, M.; Fredersdorf, S.; Griese, D.P.; Kramer, B.K.; Kromer, E.P. Experimental diabetes and left ventricular hypertrophy: Effects of beta-receptor blockade. Cardiovasc. Pathol. 2002, 11, 229–237. [Google Scholar] [CrossRef]
- Kopp, S.; Slumstrup, L.; Corydon, T.J.; Sahana, J.; Aleshcheva, G.; Islam, T.; Magnusson, N.E.; Wehland, M.; Bauer, J.; Infanger, M.; et al. Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Infanger, M.; Westphal, K.; Ulbrich, C.; Pietsch, J.; Kossmehl, P.; Vadrucci, S.; Baatout, S.; Flick, B.; Paul, M.; et al. Adelayed type of three-dimensional growth of human endothelial cells under simulated weightlessness. Tissue Eng. Part A 2009, 15, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Kossmehl, P.; Kurth, E.; Faramarzi, S.; Habighorst, B.; Shakibaei, M.; Wehland, M.; Kreutz, R.; Infanger, M.; AH, J.D.; Grosse, J.; et al. Mechanisms of apoptosis after ischemia and reperfusion: Role of the renin-angiotensin system. Apoptosis 2006, 11, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Schoenberger, J.; Bauer, J.; Moosbauer, J.; Eilles, C.; Grimm, D. Innovative strategies in in vivo apoptosis imaging. Curr. Med. Chem. 2008, 15, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Pohl, F.; Grosse, J.; Grimm, D.; Brockhoff, G.; Westphal, K.; Moosbauer, J.; Koelbl, O.; Infanger, M.; Eilles, C.; Schoenberger, J. Changes of apoptosis, p53, and BCL-2 by irradiation in poorly differentiated thyroid carcinoma cell lines: A prognostic marker for the prospect of therapeutic success? Thyroid 2010, 20, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Wehland, M.; Ma, X.; Braun, M.; Hauslage, J.; Hemmersbach, R.; Bauer, J.; Grosse, J.; Infanger, M.; Grimm, D. The impact of altered gravity and vibration on endothelial cells during a parabolic flight. Cell. Physiol. Biochem. 2013, 31, 432–451. [Google Scholar] [CrossRef] [PubMed]
- Grosse, J.; Warnke, E.; Wehland, M.; Pietsch, J.; Pohl, F.; Wise, P.; Magnusson, N.E.; Eilles, C.; Grimm, D. Mechanisms of apoptosis in irradiated and sunitinib-treated follicular thyroid cancer cells. Apoptosis 2014, 19, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Bauer, J.; Braun, M.; Schwarzwalder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, Y.; Steller, H. Live to die another way: Modes of programmed cell death and the signals emanating from dying cells. Nat. Rev. Mol. Cell Biol. 2015, 16, 329–344. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.S.; Schnellmann, R.G. Measurement of cell death in mammalian cells. Curr. Protoc. Pharmacol. 2004, 12, 12–18. [Google Scholar]
- Ashkenazi, A.; Dixit, V.M. Death receptors: Signaling and modulation. Science 1998, 281, 1305–1308. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Yonehara, S.; Ishii, A.; Yonehara, M.; Mizushima, S.; Sameshima, M.; Hase, A.; Seto, Y.; Nagata, S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 1991, 66, 233–243. [Google Scholar] [CrossRef]
- Yonehara, S.; Ishii, A.; Yonehara, M. Acell-killing monoclonal antibody (anti-Fas) to a cell surface antigen co-downregulated with the receptor of tumor necrosis factor. J. Exp. Med. 1989, 169, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.K. Death effecter domain for the assembly of death-inducing signaling complex. Apoptosis 2015, 20, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Siegmund, D.; Lang, I.; Wajant, H. Cell death-independent activities of the death receptors CD95, TRAILR1, and TRAILR2. FEBS J. 2017, 284, 1131–1159. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, Y. Role of BCL-2 family proteins in apoptosis: Apoptosomes or mitochondria? Genes Cells 1998, 3, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Haupt, S.; Berger, M.; Goldberg, Z.; Haupt, Y. Apoptosis—The p53 network. J. Cell Sci. 2003, 116, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Giaccia, A.J.; Kastan, M.B. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes Dev. 1998, 12, 2973–2983. [Google Scholar] [CrossRef] [PubMed]
- Vollmar, B.; El-Gibaly, A.M.; Scheuer, C.; Strik, M.W.; Bruch, H.P.; Menger, M.D. Acceleration of cutaneous wound healing by transient p53 inhibition. Lab. Investig. 2002, 82, 1063–1071. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; West, A.P.; Ghosh, S. NF-κB and the immune response. Oncogene 2006, 25, 6758–6780. [Google Scholar] [CrossRef] [PubMed]
- Siebenlist, U.; Franzoso, G.; Brown, K. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 1994, 10, 405–455. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.Z.; Chaturvedi, V.; Denning, M.F.; Choubey, D.; Diaz, M.O.; Nickoloff, B.J. Role of NF-κB in the apoptotic-resistant phenotype of keratinocytes. J. Biol. Chem. 1999, 274, 37957–37964. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.S.; Freiberg, R.A.; Hinata, K.; Khavari, P.A. NF-κB determines localization and features of cell death in epidermis. J. Clin. Investig. 2000, 105, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shillinglaw, W.; Henzel, W.J.; Beg, A.A. The Rela(p65) subunit of NF-κB is essential for inhibiting double-stranded RNA-induced cytotoxicity. J. Biol. Chem. 2001, 276, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Saleem, M.; Qadir, M.I.; Perveen, N.; Ahmad, B.; Saleem, U.; Irshad, T.; Ahmad, B. Inhibitors of apoptotic proteins: New targets for anticancer therapy. Chem. Biol. Drug Des. 2013, 82, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Dubrez-Daloz, L.; Dupoux, A.; Cartier, J. IAPs: More than just inhibitors of apoptosis proteins. Cell Cycle 2008, 7, 1036–1046. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Li, X.M. The IAP family: Endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000, 10, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Zangemeister-Wittke, U.; Simon, H.U. An IAP in action: The multiple roles of survivin in differentiation, immunity and malignancy. Cell Cycle 2004, 3, 1121–1123. [Google Scholar] [CrossRef] [PubMed]
- Dallaglio, K.; Marconi, A.; Pincelli, C. Survivin: A dual player in healthy and diseased skin. J. Investig. Dermatol. 2012, 132, 18–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marconi, A.; Dallaglio, K.; Lotti, R.; Vaschieri, C.; Truzzi, F.; Fantini, F.; Pincelli, C. Survivin identifies keratinocyte stem cells and is downregulated by anti-beta1 integrin during anoikis. Stem Cells 2007, 25, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Liu, T.; Cotter, M.A.; Florell, S.R.; Robinette, K.; Hanks, A.N.; Grossman, D. Melanocyte expression of survivin promotes development and metastasis of UV-induced melanoma in HGF-transgenic mice. Cancer Res. 2007, 67, 5172–5178. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, C.; Pietsch, J.; Grosse, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Hauslage, J.; Hemmersbach, R.; Braun, M.; et al. Differential gene regulation under altered gravity conditions in follicular thyroid cancer cells: Relationship between the extracellular matrix and the cytoskeleton. Cell. Physiol. Biochem. 2011, 28, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Iorio, V.; Troughton, L.D.; Hamill, K.J. Laminins: Roles and Utility in Wound Repair. Adv. Wound Care 2015, 4, 250–263. [Google Scholar] [CrossRef] [PubMed]
- Kossmehl, P.; Schonberger, J.; Shakibaei, M.; Faramarzi, S.; Kurth, E.; Habighorst, B.; von Bauer, R.; Wehland, M.; Kreutz, R.; Infanger, M.; et al. Increase of fibronectin and osteopontin in porcine hearts following ischemia and reperfusion. J. Mol. Med. 2005, 83, 626–637. [Google Scholar] [CrossRef] [PubMed]
- Infanger, M.; Shakibaei, M.; Kossmehl, P.; Hollenberg, S.M.; Grosse, J.; Faramarzi, S.; Schulze-Tanzil, G.; Paul, M.; Grimm, D. Intraluminal application of vascular endothelial growth factor enhances healing of microvascular anastomosis in a rat model. J. Vasc. Res. 2005, 42, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Infanger, M.; Grosse, J.; Westphal, K.; Leder, A.; Ulbrich, C.; Paul, M.; Grimm, D. Vascular endothelial growth factor induces extracellular matrix proteins and osteopontin in the umbilical artery. Ann. Vasc. Surg. 2008, 22, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Lund, S.A.; Giachelli, C.M.; Scatena, M. The role of osteopontin in inflammatory processes. J. Cell Commun. Signal. 2009, 3, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Dalal, S.; Zha, Q.; Daniels, C.R.; Steagall, R.J.; Joyner, W.L.; Gadeau, A.P.; Singh, M.; Singh, K. Osteopontin stimulates apoptosis in adult cardiac myocytes via the involvement of CD44 receptors, mitochondrial death pathway, and endoplasmic reticulum stress. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H1182–H1191. [Google Scholar] [CrossRef] [PubMed]
- Grosse, J.; Wehland, M.; Pietsch, J.; Schulz, H.; Saar, K.; Hubner, N.; Eilles, C.; Bauer, J.; Abou-El-Ardat, K.; Baatout, S.; et al. Gravity-sensitive signaling drives 3-dimensional formation of multicellular thyroid cancer spheroids. FASEB J. 2012, 26, 5124–5140. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P. Areagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques 1993, 15, 532–534, 536–537. [Google Scholar] [PubMed]
- Darzynkiewicz, Z.; Juan, G.; Li, X.; Gorczyca, W.; Murakami, T.; Traganos, F. Cytometry in cell necrobiology: Analysis of apoptosis and accidental cell death (necrosis). Cytometry 1997, 27, 1–20. [Google Scholar] [CrossRef]
- Simon, D.; Hoesli, S.; Roth, N.; Staedler, S.; Yousefi, S.; Simon, H.U. Eosinophil extracellular DNA traps in skin diseases. J. Allergy Clin. Immunol. 2011, 127, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Dollerup, P.; Thomsen, T.M.; Nejsum, L.N.; Faerch, M.; Osterbrand, M.; Gregersen, N.; Rittig, S.; Christensen, J.H.; Corydon, T.J. Partial nephrogenic diabetes insipidus caused by a novel AQP2 variation impairing trafficking of the aquaporin-2 water channel. BMC Nephrol. 2015, 16, 217. [Google Scholar] [CrossRef] [PubMed]
- Gehrmann, M.; Marienhagen, J.; Eichholtz-Wirth, H.; Fritz, E.; Ellwart, J.; Jaattela, M.; Zilch, T.; Multhoff, G. Dual function of membrane-bound heat shock protein 70 (Hsp70), Bag-4, and Hsp40: Protection against radiation-induced effects and target structure for natural killer cells. Cell Death Differ. 2005, 12, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [PubMed]
- Riwaldt, S.; Pietsch, J.; Sickmann, A.; Bauer, J.; Braun, M.; Segerer, J.; Schwarzwalder, A.; Aleshcheva, G.; Corydon, T.J.; Infanger, M.; et al. Identification of proteins involved in inhibition of spheroid formation under microgravity. Proteomics 2015, 15, 2945–2952. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, J.; Riwaldt, S.; Bauer, J.; Sickmann, A.; Weber, G.; Grosse, J.; Infanger, M.; Eilles, C.; Grimm, D. Interaction of proteins identified in human thyroid cells. Int. J. Mol. Sci. 2013, 14, 1164–1178. [Google Scholar] [CrossRef] [PubMed]
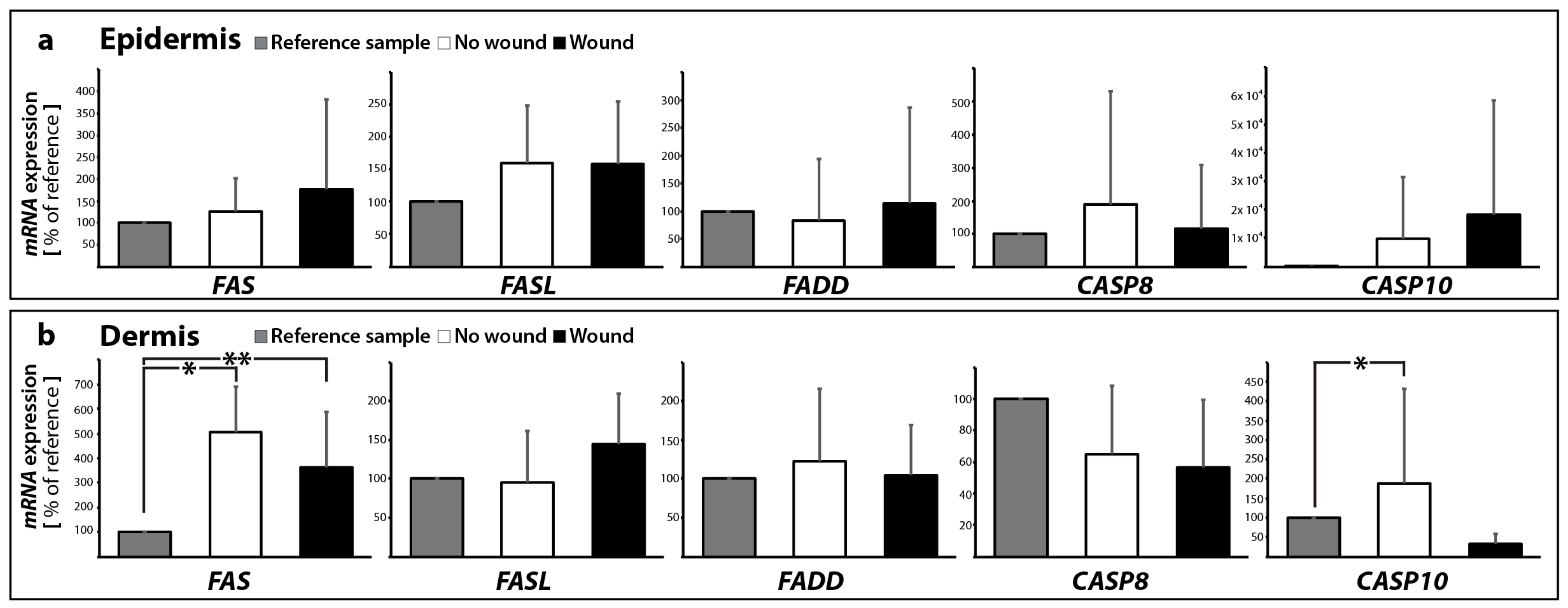
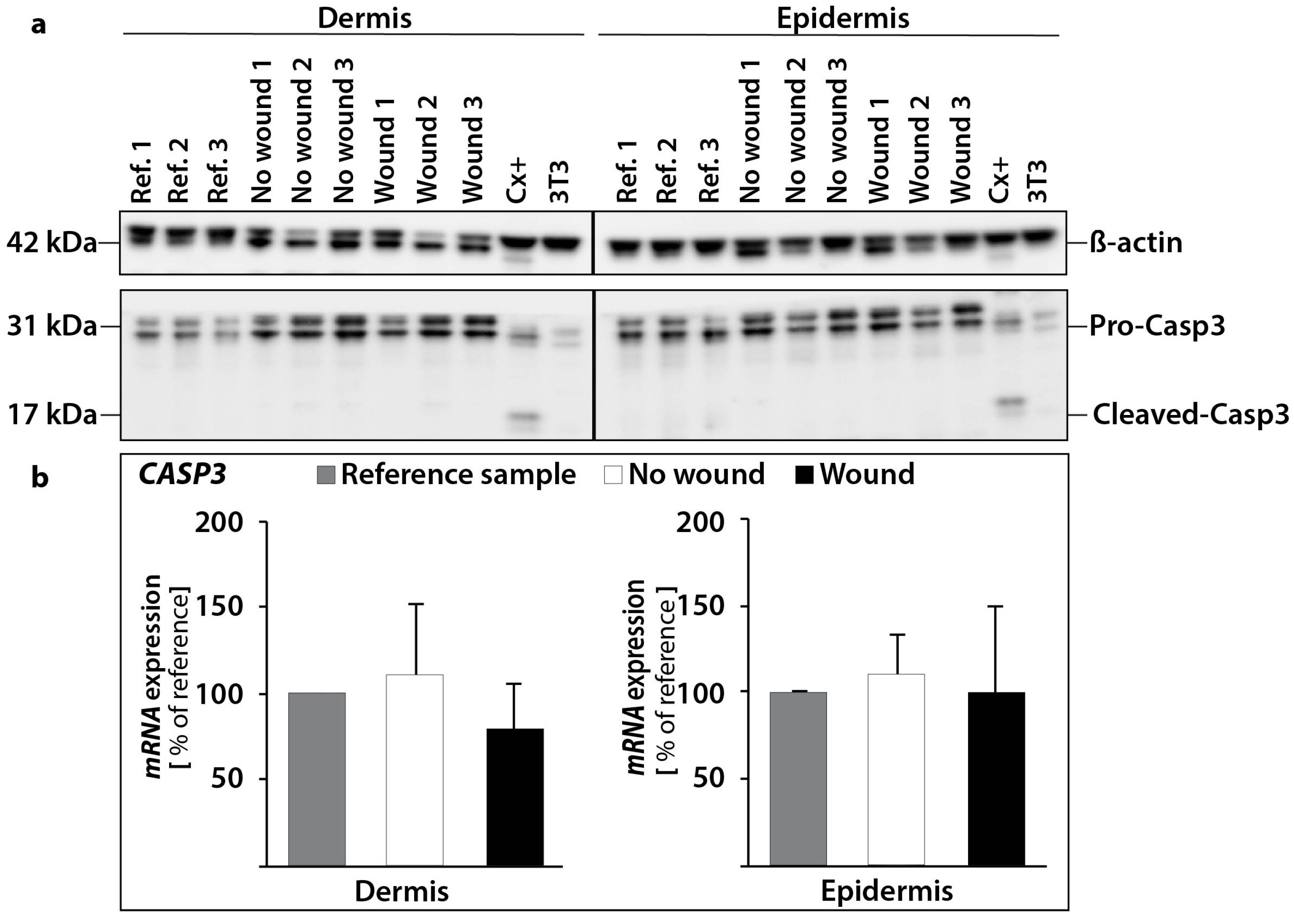

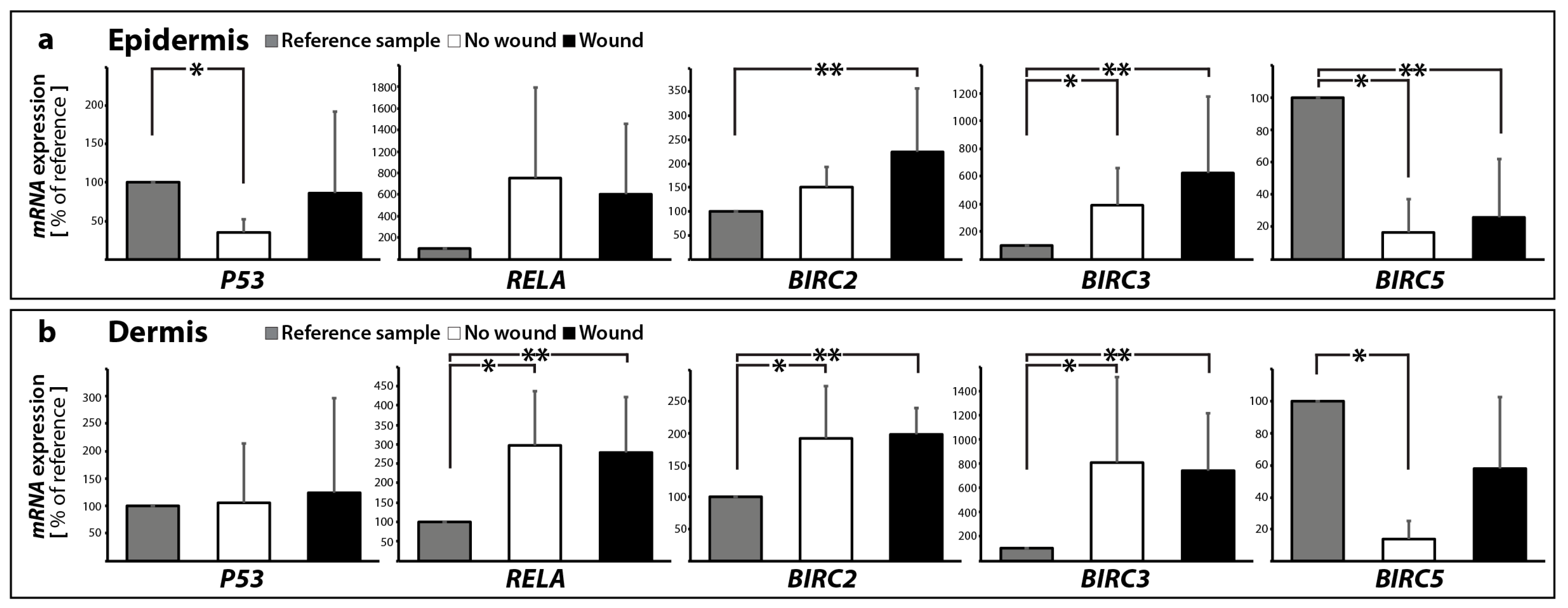
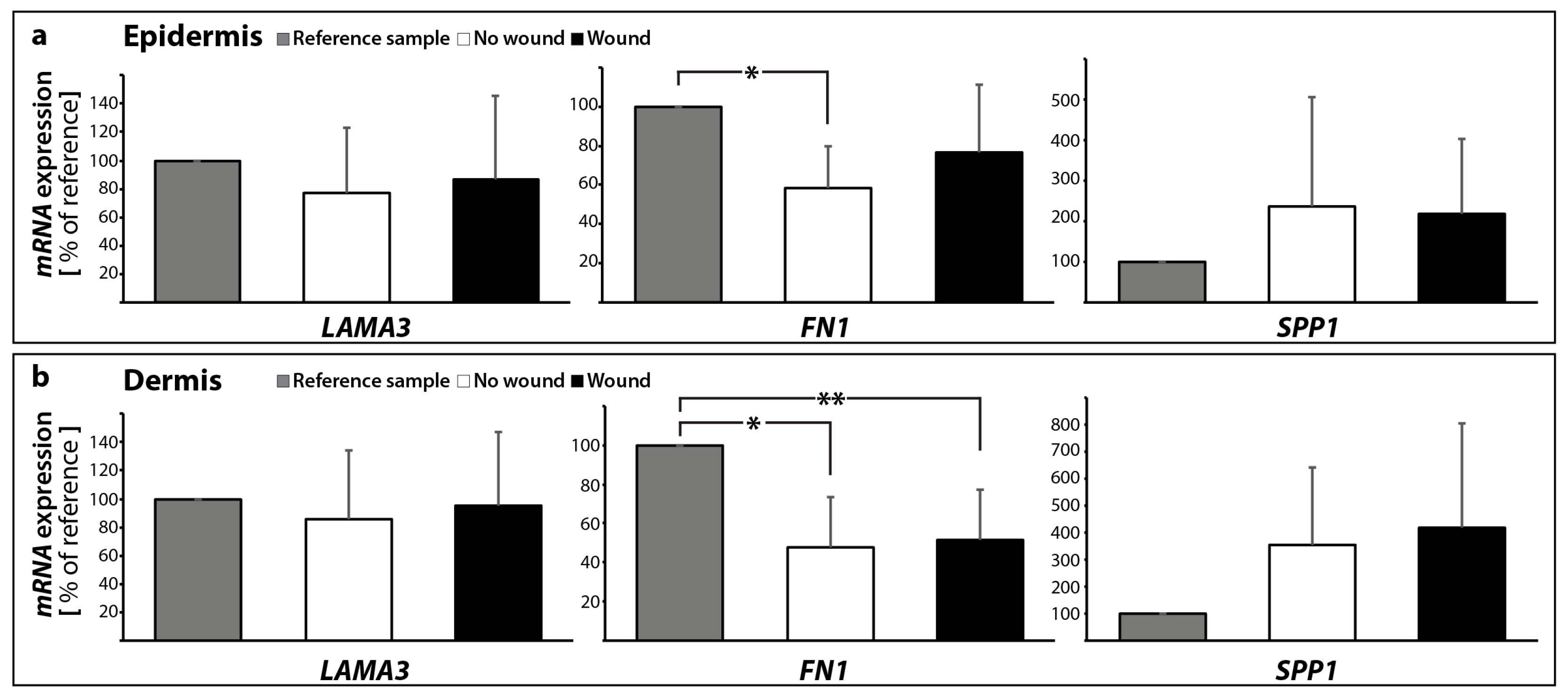
| Gene Symbol | No Wound | Wound | |||
|---|---|---|---|---|---|
| Epidermis | Dermis | Epidermis | Dermis | ||
| extrinsic pathway | FASL | 1.59 ± 0.89 | 0.96 ± 0.65 | 1.58 ± 0.97 | 1.45 ± 0.65 |
| FAS | 1.26 ± 0.76 | 5.08 ± 1.83 * | 1.78 ± 2.04 | 3.63 ± 2.24 * | |
| BIRC2 | 1.51 ± 0.42 | 1.93 ± 0.82 * | 2.25 ± 1.32 * | 1.98 ± 0.42 * | |
| BIRC3 | 3.87 ± 2.70 * | 8.10 ± 7.08 * | 6.22 ± 5.50 * | 7.41 ± 4.73 * | |
| FADD | 0.82 ± 1.12 | 1.23 ± 0.93 | 1.14 ± 1.71 | 1.05 ± 0.65 | |
| CASP8 | 1.88 ± 3.44 | 0.65 ± 0.43 | 1.18 ± 1.90 | 0.57 ± 0.43 | |
| CASP10 | 48.7 ± 107.9 | 1.89 ± 2.43 * | 90.8 ± 201.9 | 0.32 ± 0.26 | |
| CASP3 | 1.10 ± 0.23 | 1.10 ± 0.42 | 1.00 ± 0.49 | 0.80 ± 0.26 | |
| intrinsic pathway | BAX | 1.14 ± 0.49 | 1.73 ± 0.97 | 1.44 ± 1.16 | 2.41 ± 1.98 |
| BCL2 | 0.84 ± 0.48 | 1.02 ± 0.61 | 2.26 ± 2.70 | 3.23 ± 3.57 | |
| CYC1 | 1.51 ± 1.37 | 0.76 ± 0.19 | 1.79 ± 1.77 | 0.84 ± 0.26 | |
| APAF1 | 0.91 ± 0.57 | 0.81 ± 0.70 | 3.90 ± 5.81 | 7.53 ± 13.04 | |
| CASP9 | 0.30 ± 0.17 * | 0.38 ± 0.27 * | 0.32 ± 0.27 * | 0.33 ± 0.41 | |
| RELA | 7.53 ± 8.46 | 2.96 ± 1.40 * | 6.06 ± 6.50 | 2.79 ± 1.43 * | |
| BIRC5 | 0.16 ± 0.21 * | 0.14 ± 0.11 * | 0.26 ± 0.36 * | 0.58 ± 0.45 | |
| P53 | 0.35 ± 0.17 * | 1.06 ± 1.07 | 0.86 ± 1.06 | 1.24 ± 1.74 | |
| ECM | LAMA3 | 0.77 ± 0.45 | 0.86 ± 0.48 | 0.87 ± 0.58 | 0.95 ± 0.51 |
| FN1 | 0.59 ± 0.22 * | 0.48 ± 0.25 * | 0.77 ± 0.35 | 0.52 ± 0.26 * | |
| SPP1 | 2.35 ± 2.71 | 3.54 ± 2.85 | 2.18 ± 1.83 | 4.17 ± 3.88 | |
| Gene | Primer Name | Sequence |
|---|---|---|
| TBP | TATA-F | GTGACCCAGCATCACTGTTTC |
| TATA-R | GCAAACCAGAAACCCTTGCG | |
| APAF1 | APAF1-F | GGAGGATATATTAAGTGGTGGAACG |
| APAF1-R | GTTTTGAAGTCAGGGGACACG | |
| BAX | BAX-F | GTCAGCTGCCACTCGGAAA |
| BAX-R | AGTAACATGGAGCTGCAGAGGAT | |
| BCL2 | BCL2-F | CCTGTGGATGACTGAGTACCTGAA |
| BCL2-R | TCAGAGACAGCCAGGAGAAATCA | |
| BIRC2 | BIRC2-F | GCTTTTGTTGTGATGGTGGCT |
| BIRC2-R | ACTCACACCTTGGAAACCACT | |
| BIRC3 | BIRC3-F | TGCTGTGATGGTGGACTCAG |
| BIRC3-R | ACTCACACCTTGGAAACCACT | |
| BIRC5 | BIRC5-F | GCCAGATGACGACCCCATAG |
| BIRC5-R | CACCAAGGGTTAATTCTTCAAACTG | |
| CASP3 | CASP3-F | AACTGCTCCTTTTGCTGTGATCT |
| CASP3-R | GCAGCAAACCTCAGGGAAAC | |
| CASP8 | CASP8-F | TGCAAAAGCACGGGAGAAAG |
| CASP8-R | CTCTTCAAAGGTCGTGGTCAAAG | |
| CASP9 | CASP9-F | CTCCAACATCGACTGTGAGAAGTT |
| CASP9-R | GCGCCAGCTCCAGCAA | |
| CASP10 | CASP10-F | GACAAGGAAGCCGAGTCGTA |
| CASP10-R | GCCTCTGTGGTTCCGATTCA | |
| CYC1 | CYC1-F | CACTGCGGGAAGGTCTCTAC |
| CYC1-R | GGGGTGCCATCGTCAAACTC | |
| FADD | FADD-F | CCTGGGGAAGAAGACCTGTGTG |
| FADD-R | TCGATGCTGTCGATCTTGGTG | |
| FAS | FAS-F | AGTCTGGTTCATCCCCATTGAC |
| FAS-R | AGGGATTGGAATTGAGGAAGACT | |
| FASL | FASL-F | CTTCACTCCAGAAAGCAGGACAAT |
| FASL-R | AGGGATTGGAATTGAGGAAGACT | |
| FN1 | FN1-F | AGATCTACCTGTACACCTTGAATGACA |
| FN1-R | CATGATACCAGCAAGGAATTGG | |
| LAMA3 | LAMA3-F | AAAGCAAGAAGTCAGTCCAGC |
| LAMA3-R | TCCCATGAAGACCATCTCGG | |
| P53 | P53-F | CCTGGATTGGCCAGACTGC |
| P53-R | TTTTCAGGAAGTAGTTTCCATAGGT | |
| RELA | RELA-F | CGCTTCTTCACACACTGGATTC |
| RELA-R | ACTGCCGGGATGGCTTCT | |
| SPP1 | SPP1-F | CGAGGTGATAGTGTGGTTTATGGA |
| SPP1-R | CGTCTGTAGCATCAGGGTACTG |
| Primary Antibody | Company | Dilution | Molecular Weight (kDa) | Catalogue Number |
|---|---|---|---|---|
| Caspase-3 | Abcam | 1:1000 | 31, 21–14 | Ab13585 |
| β-actin | Sigma | 1:1000 | 42 | A5316 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riwaldt, S.; Monici, M.; Graver Petersen, A.; Birk Jensen, U.; Evert, K.; Pantalone, D.; Utpatel, K.; Evert, M.; Wehland, M.; Krüger, M.; et al. Preparation of A Spaceflight: Apoptosis Search in Sutured Wound Healing Models. Int. J. Mol. Sci. 2017, 18, 2604. https://doi.org/10.3390/ijms18122604
Riwaldt S, Monici M, Graver Petersen A, Birk Jensen U, Evert K, Pantalone D, Utpatel K, Evert M, Wehland M, Krüger M, et al. Preparation of A Spaceflight: Apoptosis Search in Sutured Wound Healing Models. International Journal of Molecular Sciences. 2017; 18(12):2604. https://doi.org/10.3390/ijms18122604
Chicago/Turabian StyleRiwaldt, Stefan, Monica Monici, Asbjørn Graver Petersen, Uffe Birk Jensen, Katja Evert, Desiré Pantalone, Kirsten Utpatel, Matthias Evert, Markus Wehland, Marcus Krüger, and et al. 2017. "Preparation of A Spaceflight: Apoptosis Search in Sutured Wound Healing Models" International Journal of Molecular Sciences 18, no. 12: 2604. https://doi.org/10.3390/ijms18122604







