Immunoinformatics Features Linked to Leishmania Vaccine Development: Data Integration of Experimental and In Silico Studies
Abstract
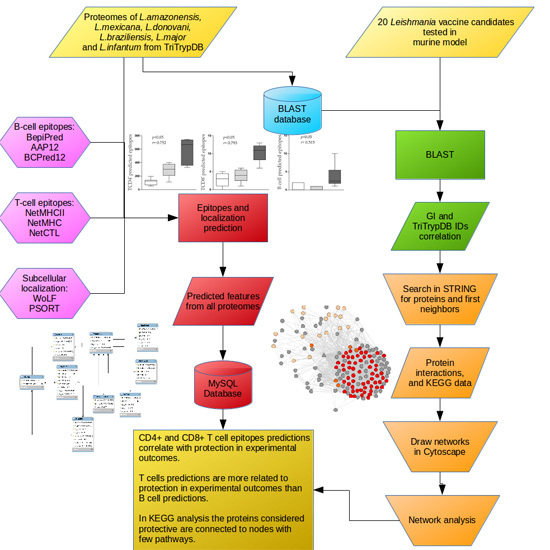
:1. Introduction
2. Results
2.1. Leishmania Proteins Dataset Selection
2.2. Epitope and Subcellular Localization Predictions
2.3. Predicted Epitopes and Experimental Outcome Correlation
2.4. Number of Alleles (NA) and Experimental Outcome (EO) Correlation
2.5. Mapping Immunogenic Proteins on Protein-Protein Interaction Networks (PPI Networks)
3. Discussion
4. Materials and Methods
4.1. Selection of the Leishmania Antigens
4.2. Leishmania Proteome Data
4.3. Epitope and Subcellular Localization Predictions
4.4. Development of Relational Database
4.5. Mapping Immunogenic Proteins on Protein-Protein Interaction Networks (PPI Networks)
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Peacock, C.S.; Seeger, K.; Harris, D.; Murphy, L.; Ruiz, J.C.; Quail, M.A.; Peters, N.; Adlem, E.; Tivey, A.; Aslett, M.; et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007, 39, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, P.M.; Beaumier, C.M.; Strych, U.; Hayward, T.; Hotez, P.J.; Bottazzi, M.E. Status of vaccine research and development of vaccines for leishmaniasis. Vaccine 2016, 34, 2992–2995. [Google Scholar] [CrossRef] [PubMed]
- Rezende, A.M.; Folador, E.L.; Resende, D.M.; Ruiz, J.C. Computational prediction of protein–protein interactions in Leishmania predicted proteomes. PLoS ONE 2012, 7, e51304. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ye, B.; Han, Z.; Huang, M.; Zhu, Y. Comparison of transcriptional profiles between CD4+ and CD8+ T cells in HIV type 1-infected patients. AIDS Res. Hum. Retrovir. 2014, 30, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Salay, G.; Dorta, M.L.; Santos, N.M.; Mortara, R.A.; Brodskyn, C.; Oliveira, C.I.; Barbieri, C.L.; Rodrigues, M.M. Testing of four Leishmania vaccine candidates in a mouse model of infection with Leishmania (Viannia) braziliensis, the main causative agent of cutaneous leishmaniasis in the New World. Clin. Vaccine Immunol. CVI 2007, 14, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Diao, H.; Ji, J.; Soong, L. DNA immunization with the gene encoding P4 nuclease of Leishmania amazonensis protects mice against cutaneous Leishmaniasis. Infect. Immun. 2003, 71, 6270–6278. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, C.E.; Ferreira, J.H.; Mussalem, J.S.; Longo-Maugeri, I.M.; Gentil, L.G.; dos Santos, M.R.; Katz, S.; Barbieri, C.L. Partial protective responses induced by a recombinant cysteine proteinase from Leishmania (Leishmania) amazonensis in a murine model of cutaneous leishmaniasis. Exp. Parasitol. 2010, 124, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Montalvo-Alvarez, A.M.; Folgueira, C.; Carrion, J.; Monzote-Fidalgo, L.; Canavate, C.; Requena, J.M. The Leishmania HSP20 is antigenic during natural infections, but, as DNA vaccine, it does not protect BALB/c mice against experimental L. amazonensis infection. J. Biomed. Biotechnol. 2008, 2008, 695432. [Google Scholar] [CrossRef] [PubMed]
- Champsi, J.; McMahon-Pratt, D. Membrane glycoprotein M-2 protects against Leishmania amazonensis infection. Infect. Immun. 1988, 56, 3272–3279. [Google Scholar] [PubMed]
- Gonzalez, C.R.; Noriega, F.R.; Huerta, S.; Santiago, A.; Vega, M.; Paniagua, J.; Ortiz-Navarrete, V.; Isibasi, A.; Levine, M.M. Immunogenicity of a Salmonella typhi CVD 908 candidate vaccine strain expressing the major surface protein gp63 of Leishmania mexicana mexicana. Vaccine 1998, 16, 1043–1052. [Google Scholar] [CrossRef]
- Mendez, S.; Gurunathan, S.; Kamhawi, S.; Belkaid, Y.; Moga, M.A.; Skeiky, Y.A.; Campos-Neto, A.; Reed, S.; Seder, R.A.; Sacks, D. The potency and durability of DNA- and protein-based vaccines against Leishmania major evaluated using low-dose, intradermal challenge. J. Immunol. 2001, 166, 5122–5128. [Google Scholar] [CrossRef] [PubMed]
- Rivier, D.; Bovay, P.; Shah, R.; Didisheim, S.; Mauel, J. Vaccination against Leishmania major in a CBA mouse model of infection: Role of adjuvants and mechanism of protection. Parasite Immunol. 1999, 21, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Sjolander, A.; Baldwin, T.M.; Curtis, J.M.; Bengtsson, K.L.; Handman, E. Vaccination with recombinant Parasite Surface Antigen 2 from Leishmania major induces a Th1 type of immune response but does not protect against infection. Vaccine 1998, 16, 2077–2084. [Google Scholar] [CrossRef]
- Webb, J.R.; Campos-Neto, A.; Ovendale, P.J.; Martin, T.I.; Stromberg, E.J.; Badaro, R.; Reed, S.G. Human and murine immune responses to a novel Leishmania major recombinant protein encoded by members of a multicopy gene family. Infect. Immun. 1998, 66, 3279–3289. [Google Scholar] [PubMed]
- Solioz, N.; Blum-Tirouvanziam, U.; Jacquet, R.; Rafati, S.; Corradin, G.; Mauel, J.; Fasel, N. The protective capacities of histone H1 against experimental murine cutaneous leishmaniasis. Vaccine 1999, 18, 850–859. [Google Scholar] [CrossRef]
- Soussi, N.; Milon, G.; Colle, J.H.; Mougneau, E.; Glaichenhaus, N.; Goossens, P.L. Listeria monocytogenes as a short-lived delivery system for the induction of type 1 cell-mediated immunity against the p36/LACK antigen of Leishmania major. Infect. Immun. 2000, 68, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Carrion, J.; Folgueira, C.; Alonso, C. Immunization strategies against visceral leishmaniosis with the nucleosomal histones of Leishmania infantum encoded in DNA vaccine or pulsed in dendritic cells. Vaccine 2008, 26, 2537–2544. [Google Scholar] [CrossRef] [PubMed]
- Santos-Gomes, G.M.; Rodrigues, A.; Teixeira, F.; Carreira, J.; Alexandre-Pires, G.; Carvalho, S.; Santos-Mateus, D.; Martins, C.; Vale-Gato, I.; Marques, C.; et al. Immunization with the Leishmania infantum recombinant cyclophilin protein 1 confers partial protection to subsequent parasite infection and generates specific memory T cells. Vaccine 2014, 32, 1247–1253. [Google Scholar] [CrossRef] [PubMed]
- Agallou, M.; Smirlis, D.; Soteriadou, K.P.; Karagouni, E. Vaccination with Leishmania histone H1-pulsed dendritic cells confers protection in murine visceral leishmaniasis. Vaccine 2012, 30, 5086–5093. [Google Scholar] [CrossRef] [PubMed]
- Khoshgoo, N.; Zahedifard, F.; Azizi, H.; Taslimi, Y.; Alonso, M.J.; Rafati, S. Cysteine proteinase type III is protective against Leishmania infantum infection in BALB/c mice and highly antigenic in visceral leishmaniasis individuals. Vaccine 2008, 26, 5822–5829. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Be, I.; da Silva Zardo, R.; Paraguai de Souza, E.; Borja-Cabrera, G.P.; Rosado-Vallado, M.; Mut-Martin, M.; Garcia-Miss Mdel, R.; Palatnik de Sousa, C.B.; Dumonteil, E. Cross-protective efficacy of a prophylactic Leishmania donovani DNA vaccine against visceral and cutaneous murine leishmaniasis. Infect. Immun. 2005, 73, 812–819. [Google Scholar] [CrossRef]
- Ghosh, A.; Zhang, W.W.; Matlashewski, G. Immunization with A2 protein results in a mixed Th1/Th2 and a humoral response which protects mice against Leishmania donovani infections. Vaccine 2001, 20, 59–66. [Google Scholar] [CrossRef]
- Resende, D.M.; Rezende, A.M.; Oliveira, N.J.; Batista, I.C.; Correa-Oliveira, R.; Reis, A.B.; Ruiz, J.C. An assessment on epitope prediction methods for protozoa genomes. BMC Bioinform. 2012, 13, 309. [Google Scholar] [CrossRef] [PubMed]
- Costa, C.H.; Peters, N.C.; Maruyama, S.R.; de Brito, E.C., Jr.; Santos, I.K. Vaccines for the leishmaniases: Proposals for a research agenda. PLoS Negl. Trop. Dis. 2011, 5, e943. [Google Scholar]
- Reed, S.G.; Coler, R.N.; Campos-Neto, A. Development of a leishmaniasis vaccine: The importance of MPL. Expert Rev. Vaccines 2003, 2, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Lundegaard, C.; Lamberth, K.; Buus, S.; Lund, O.; Nielsen, M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 2007, 8, 424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, M.; Lundegaard, C.; Lund, O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinform. 2007, 8, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsen, J.E.; Lund, O.; Nielsen, M. Improved method for predicting linear B-cell epitopes. Immun. Res. 2006, 2, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Liu, H.; Yang, J.; Chou, K.C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 2007, 33, 423–428. [Google Scholar] [CrossRef] [PubMed]
- El Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. 2008, 21, 243–255. [Google Scholar] [CrossRef] [PubMed]
- El Manzalawy, Y.; Dobbs, D.; Honavar, V. Predicting flexible length linear B-cell epitopes. Comput. Syst. Bioinform. Conf. 2008, 7, 121–132. [Google Scholar]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Najera, C.; Pina-Aguilar, R.; Xacur-Garcia, F.; Ramirez-Sierra, M.J.; Dumonteil, E. Mining the Leishmania genome for novel antigens and vaccine candidates. Proteomics 2009, 9, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Agallou, M.; Athanasiou, E.; Koutsoni, O.; Dotsika, E.; Karagouni, E. Experimental Validation of Multi-Epitope Peptides Including Promising MHC Class I- and II-Restricted Epitopes of Four Known Leishmania infantum Proteins. Front. Immunol. 2014, 5, 268. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Queiroz, A.T.; Tosta, R.; Carvalho, A.M.; Barbosa, C.H.; Bellio, M.; de Oliveira, C.I.; Barral-Netto, M. Prediction of CD8+ Epitopes in Leishmania braziliensis Proteins Using EPIBOT: In Silico Search and In Vivo Validation. PLoS ONE 2015, 10, e0124786. [Google Scholar] [CrossRef] [PubMed]
- Freitas e Silva, R.; Ferreira, L.F.; Hernandes, M.Z.; de Brito, M.E.; de Oliveira, B.C.; da Silva, A.A.; de-Melo-Neto, O.P.; Rezende, A.M.; Pereira, V.R. Combination of In Silico Methods in the Search for Potential CD4(+) and CD8(+) T Cell Epitopes in the Proteome of Leishmania braziliensis. Front. Immunol. 2016, 7, 327. [Google Scholar]
- Day, M.J. Immunoglobulin G subclass distribution in canine leishmaniosis: A review and analysis of pitfalls in interpretation. Vet. Parasitol. 2007, 147, 2–8. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Silva, L.; Romero, H.D.; Prata, A.; Costa, R.T.; Nascimento, E.; Carvalho, S.F.; Rodrigues, V. Immunologic tests in patients after clinical cure of visceral leishmaniasis. Am. J. Trop. Med. Hyg. 2006, 75, 739–743. [Google Scholar] [PubMed]
- Rosa, R.; Marques, C.; Rodrigues, O.R.; Santos-Gomes, G.M. Immunization with Leishmania infantum released proteins confers partial protection against parasite infection with a predominant Th1 specific immune response. Vaccine 2007, 25, 4525–4532. [Google Scholar] [CrossRef] [PubMed]
- Lemesre, J.L.; Holzmuller, P.; Goncalves, R.B.; Bourdoiseau, G.; Hugnet, C.; Cavaleyra, M.; Papierok, G. Long-lasting protection against canine visceral leishmaniasis using the LiESAp-MDP vaccine in endemic areas of France: Double-blind randomised efficacy field trial. Vaccine 2007, 25, 4223–4234. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, P.J.; Alonso, A.; Sanchez-Gorostiaga, A.; Moreno-Paz, M.; Gomez, M.J.; Ramos, I.; Parro, V.; Larraga, V. Genome-wide analysis reveals increased levels of transcripts related with infectivity in peanut lectin non-agglutinated promastigotes of Leishmania infantum. Genomics 2009, 93, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Benitez, D.; Medeiros, A.; Fiestas, L.; Panozzo-Zenere, E.A.; Maiwald, F.; Prousis, K.C.; Roussaki, M.; Calogeropoulou, T.; Detsi, A.; Jaeger, T.; et al. Identification of Novel Chemical Scaffolds Inhibiting Trypanothione Synthetase from Pathogenic Trypanosomatids. PLoS Negl. Trop. Dis. 2016, 10, e0004617. [Google Scholar] [CrossRef] [PubMed]
- Iborra, S.; Parody, N.; Abanades, D.R.; Bonay, P.; Prates, D.; Novais, F.O.; Barral-Netto, M.; Alonso, C.; Soto, M. Vaccination with the Leishmania major ribosomal proteins plus CpG oligodeoxynucleotides induces protection against experimental cutaneous leishmaniasis in mice. Microb. Inf. Inst. Pasteur 2008, 10, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Cordeiro-Da-Silva, A.; Borges, M.C.; Guilvard, E.; Ouaissi, A. Dual role of the Leishmania major ribosomal protein S3a homologue in regulation of T- and B-cell activation. Infect. Immun. 2001, 69, 6588–6596. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.; Requena, J.M.; Garcia, M.; Gomez, L.C.; Navarrete, I.; Alonso, C. Genomic organization and expression of two independent gene arrays coding for two antigenic acidic ribosomal proteins of Leishmania. J. Biol. Chem. 1993, 268, 21835–21843. [Google Scholar] [PubMed]
- Soto, M.; Alonso, C.; Requena, J.M. The Leishmania infantum acidic ribosomal protein LiP2a induces a prominent humoral response in vivo and stimulates cell proliferation in vitro and interferon-gamma (IFN-γ) production by murine splenocytes. Clin. Exp. Immunol. 2000, 122, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Chan, S.K.; Robinson, D.P.; Dwyer, D.M.; Nandan, D.; Foster, L.J.; Reiner, N.E. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008, 9, R35. [Google Scholar] [CrossRef] [PubMed]
- McConville, M.J.; Naderer, T. Metabolic pathways required for the intracellular survival of Leishmania. Annu. Rev. Microbiol. 2011, 65, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Naderer, T.; Dandash, O.; McConville, M.J. Calcineurin is required for Leishmania major stress response pathways and for virulence in the mammalian host. Mol. Microbiol. 2011, 80, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Madeira da Silva, L.; Beverley, S.M. Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc. Natl. Acad. Sci. USA 2010, 107, 11965–11970. [Google Scholar] [CrossRef] [PubMed]
- Carter, N.S.; Yates, P.; Arendt, C.S.; Boitz, J.M.; Ullman, B. Purine and pyrimidine metabolism in Leishmania. Adv. Exp. Med. Biol. 2008, 625, 141–154. [Google Scholar] [PubMed]
- Real, F.; Vidal, R.O.; Carazzolle, M.F.; Mondego, J.M.; Costa, G.G.; Herai, R.H.; Wurtele, M.; de Carvalho, L.M.; Carmona e Ferreira, R.; Mortara, R.A.; et al. The genome sequence of Leishmania (Leishmania) amazonensis: Functional annotation and extended analysis of gene models. DNA Res. 2013, 20, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.; Bulik, S.; Tampe, R.; Van Endert, P.M.; Holzhutter, H.G. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J. Immunol. 2003, 171, 1741–1749. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Lund, O.; Kesmir, C. The role of the proteasome in generating cytotoxic T-cell epitopes: Insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 2005, 57, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Buus, S.; Lauemoller, S.L.; Worning, P.; Kesmir, C.; Frimurer, T.; Corbet, S.; Fomsgaard, A.; Hilden, J.; Holm, A.; Brunak, S. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens 2003, 62, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Worning, P.; Hvid, C.S.; Lamberth, K.; Buus, S.; Brunak, S.; Lund, O. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics 2004, 20, 1388–1397. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lundegaard, C.; Worning, P.; Lauemoller, S.L.; Lamberth, K.; Buus, S.; Brunak, S.; Lund, O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003, 12, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.; Lund, O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinform. 2009, 10, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M. KEGG Bioinformatics Resource for Plant Genomics and Metabolomics. Methods Mol. Biol. 2016, 1374, 55–70. [Google Scholar] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]





| Leishmania Tropism | Geographic Area | Specie | Candidate Antigen | Function | MySQL ID | NCBI Sequence Acession | Animal | Experimental Outcome Indicative | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Dermatotropic Leishmania species | New world | L. brazilinensis | Thiol-specific-antioxidant (TSA) | Tryparedoxin peroxidase | LbrM.15.1080 | gi 154334618 | mice | No protection | [5] |
| LeiF | Leishmania putative eukaryotic initiation factor | LbrM.25.0580 | gi 154338682 | mice | No protection | [5] | |||
| LACK | Leishmania homolog of receptors for activated C-kinase | LbrM.28.2950 | gi 154340729 | mice | Partial protection | [5] | |||
| L. amazonensis | P4 nuclease partial | Endonuclease activity | A16600 | gi 29165287 | mice | No protection | [6] | ||
| Cysteine proteinase | Cysteine-type peptidase activity | A22180 | gi 30142572 | mice | Partial protection | [7] | |||
| HSP20 | Heat shock protein | A38570 | gi 513044555 | mice | No protection | [8] | |||
| GP46 | Membrane glycoprotein | A64110 | gi 159321 | mice | Protection | [9] | |||
| L. mexicana | GP63 | Metalloendopeptidase activity | LmxM.10.0465 | gi 401416782 | mice | Protection | [10] | ||
| Old world | L. major | LmTSI | Stress-induced protein sti1 | LmjF.08.1110 | gi 68124434 | mice | Protection | [11] | |
| GP63 | Metalloendopeptidase activity | LmjF.10.0470 | gi 157865341 | mice | Protection | [12] | |||
| PSA 2 | Promastigote surface antigen protein 2 | LmjF.12.1000 | gi 68124979 | mice | No protection | [13] | |||
| TSA | Thiol-specific-antioxidant—Tryparedoxin peroxidase | LmjF.15.1080 | gi 68125473 | mice | No protection | [14] | |||
| Histone H1 | DNA binding | LmjF.27.1190 | gi 4008565 | mice | No protection | [15] | |||
| LACK | Leishmania homolog of receptors for activated C-kinase | LmjF.28.2740 | gi 157872022 | mice | Partial protection | [16] | |||
| Viscerotropic Leishmania species | New world | L. infantum | H2A | DNA binding | LinJ.21.1160 | gi 339898105 | mice | No protection | [17] |
| LiCY1 | Peptidylprolyl isomerase | LinJ.25.0940 | gi 146088699 | mice | Partial protection | [18] | |||
| Histone H1 | DNA binding | LinJ.27.1070 | gi 78146500 | mice | No protection | [19] | |||
| CPC | Cysteine-type peptidase activity | LinJ.29.0860 | gi 146092987 | mice | Protection | [20] | |||
| Old world | L. donovani | NH36 | Hydrolase activity | LdBPK_181570.1 | gi 19697561 | mice | Partial protection | [21] | |
| A2 | Amastigote-specific protein—stress response protein | LdBPK_220560.1 | gi 12382244 | mice | Protection | [22] |
| MySQL ID | Prediction of Binding MHC Epitopes | Prediction of B Cells Epitopes | EO 1 | Prediction of Subcelular Localization | ||||
|---|---|---|---|---|---|---|---|---|
| Binding MHC Class I Epitopes | Binding MHC Class II Epitopes | AAP12 | BCPred12 | BepiPred | ||||
| NetMHC | NetCTL | NetMHCII | ||||||
| LbrM.15.1080 | 7 | 74 | 132 | 97 | 32 | 1 | No protection | cyt |
| LbrM.25.0580 | 6 | 31 | 121 | 75 | 23 | 2 | No protection | cyt |
| LbrM.28.2950 | 14 | 67 | 298 | 130 | 15 | 2 | Partial protection | nuc |
| A16600 | 4 | 18 | 63 | 21 | 9 | 2 | No protection | cyt |
| A22180 | 15 | 105 | 403 | 214 | 65 | 14 | Partial protection | ext |
| A38570 | 10 | 46 | 146 | 52 | 0 | 5 | No protection | ext |
| A64110 | 28 | 149 | 739 | 193 | 20 | 10 | Protection | ext |
| LmxM.10.0465 | 31 | 196 | 747 | 302 | 91 | 36 | Protection | ext |
| LmjF.08.1110 | 29 | 177 | 369 | 291 | 79 | 16 | Protection | cyt |
| LmjF.10.0470 | 27 | 177 | 668 | 317 | 52 | 19 | Protection | pla |
| LmjF.12.1000 | 23 | 100 | 475 | 226 | 102 | 12 | No protection | ext |
| LmjF.15.1080 | 9 | 77 | 199 | 81 | 35 | 5 | No protection | cyt |
| LmjF.27.1190 | 1 | 27 | 89 | 20 | 20 | 2 | No protection | nuc |
| LmjF.28.2740 | 16 | 64 | 301 | 172 | 18 | 7 | Partial protection | nuc |
| LinJ.21.1160 | 5 | 52 | 130 | 33 | 30 | 2 | No protection | nuc |
| LinJ.25.0940 | 8 | 26 | 116 | 98 | 69 | 2 | Partial protection | cyt |
| LinJ.27.1070 | 1 | 36 | 53 | 80 | 58 | 2 | No protection | nuc |
| LinJ.29.0860 | 21 | 99 | 331 | 201 | 85 | 4 | Protection | ext |
| LdBPK_181570.1 | 14 | 89 | 356 | 161 | 63 | 2 | Partial protection | ext |
| LdBPK_220560.1 | 35 | 159 | 669 | 200 | 165 | 12 | Protection | pla |
| Leishmania Specie | Version of Proteome | Predicted Proteins |
|---|---|---|
| L. braziliensis | 3.1 | 8357 |
| L. amazonensis | - 1 | 8168 |
| L. mexicana | 9.0 | 8250 |
| L. major | 9.0 | 8400 |
| L. donovani | 8.0 | 8083 |
| L. infantum | 3.2 | 8241 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brito, R.C.F.; Guimarães, F.G.; Velloso, J.P.L.; Corrêa-Oliveira, R.; Ruiz, J.C.; Reis, A.B.; Resende, D.M. Immunoinformatics Features Linked to Leishmania Vaccine Development: Data Integration of Experimental and In Silico Studies. Int. J. Mol. Sci. 2017, 18, 371. https://doi.org/10.3390/ijms18020371
Brito RCF, Guimarães FG, Velloso JPL, Corrêa-Oliveira R, Ruiz JC, Reis AB, Resende DM. Immunoinformatics Features Linked to Leishmania Vaccine Development: Data Integration of Experimental and In Silico Studies. International Journal of Molecular Sciences. 2017; 18(2):371. https://doi.org/10.3390/ijms18020371
Chicago/Turabian StyleBrito, Rory C. F., Frederico G. Guimarães, João P. L. Velloso, Rodrigo Corrêa-Oliveira, Jeronimo C. Ruiz, Alexandre B. Reis, and Daniela M. Resende. 2017. "Immunoinformatics Features Linked to Leishmania Vaccine Development: Data Integration of Experimental and In Silico Studies" International Journal of Molecular Sciences 18, no. 2: 371. https://doi.org/10.3390/ijms18020371
APA StyleBrito, R. C. F., Guimarães, F. G., Velloso, J. P. L., Corrêa-Oliveira, R., Ruiz, J. C., Reis, A. B., & Resende, D. M. (2017). Immunoinformatics Features Linked to Leishmania Vaccine Development: Data Integration of Experimental and In Silico Studies. International Journal of Molecular Sciences, 18(2), 371. https://doi.org/10.3390/ijms18020371