NANOG Expression as a Responsive Biomarker during Treatment with Hedgehog Signal Inhibitor in Acute Myeloid Leukemia
Abstract
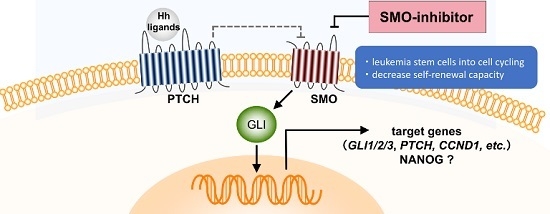
:1. Introduction
2. Results
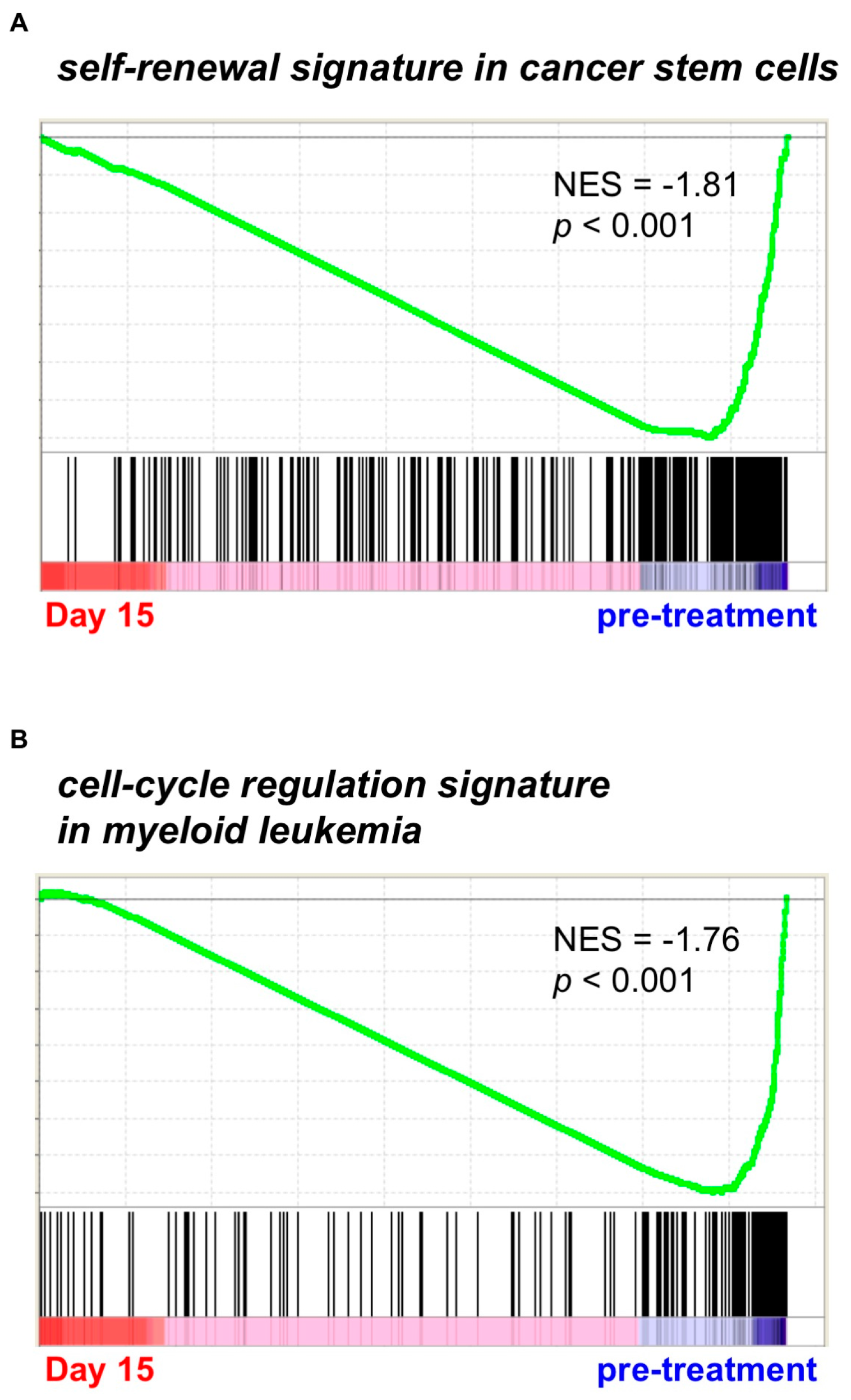
2.1. Gene Expression Profiling from AML Patients during Treatment with PF-913 Monotherapy
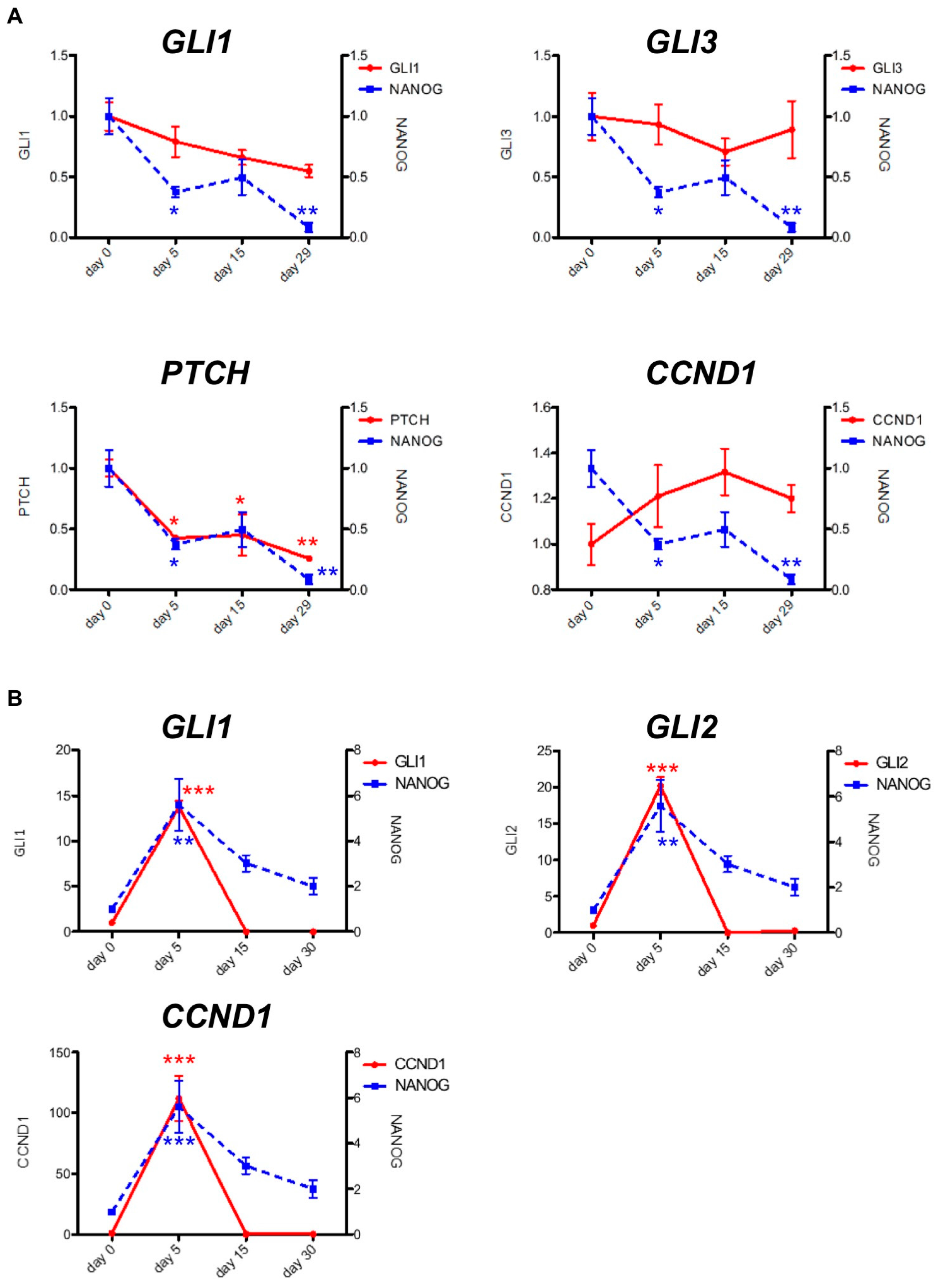
2.2. PF-913 Treatment Dysregulate NANOG Expression In Vivo
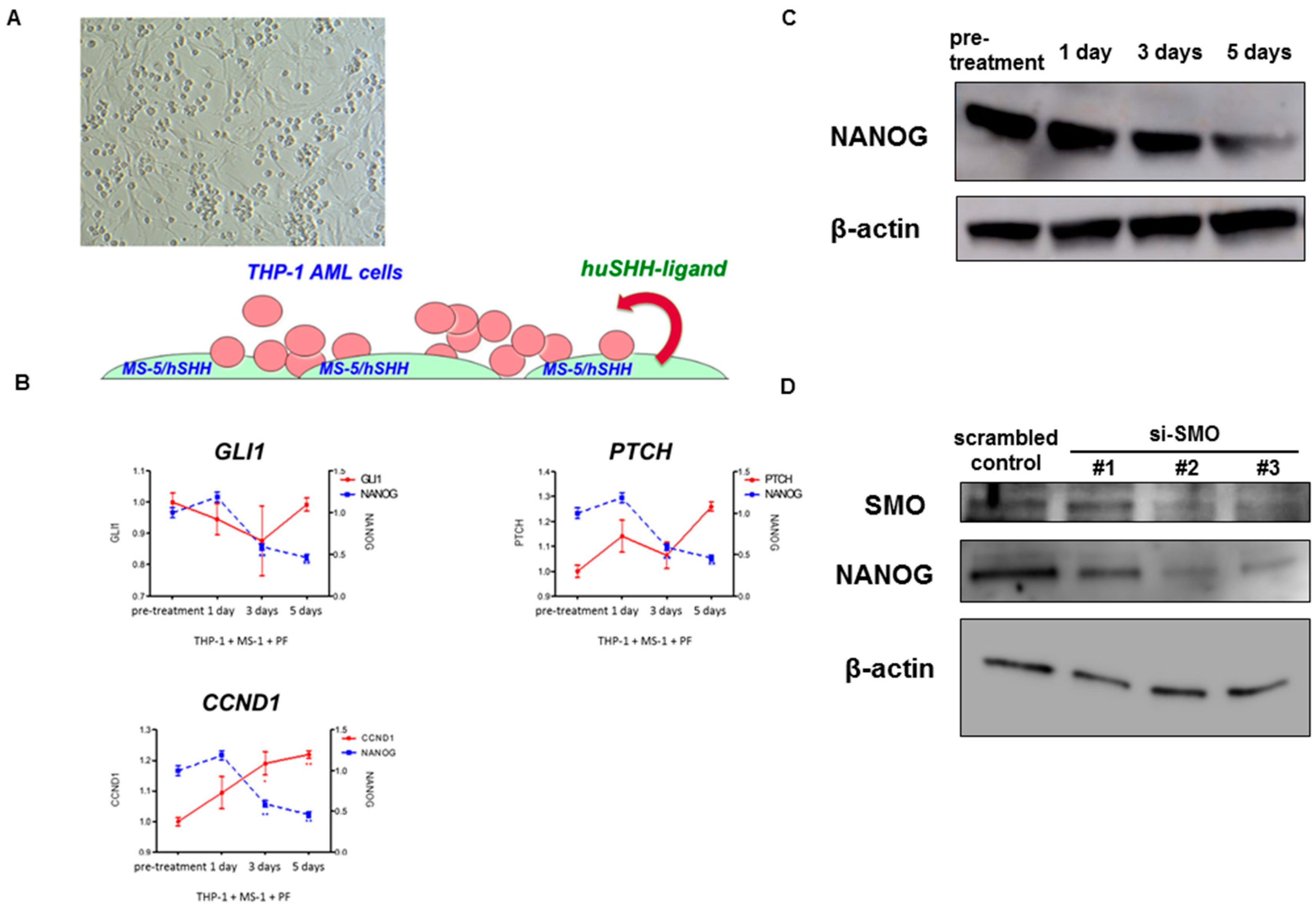
2.3. Pharmacological and Genetic Knockdown of SMO Lead to Decrease NANOG Expression In Vitro Model
3. Materials and Methods
3.1. DNA Microarray and Data Analysis
3.2. Gene Set Enrichment Analysis
3.3. Real-Time RT-PCR
3.4. Statistical Analysis
3.5. Reagents
3.6. Cells
3.7. Western Blotting and Immunohistochemistry
3.8. siRNA Transfection
3.9. Patient Characteristics
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Amakye, D.; Jagani, Z.; Dorsch, M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nat. Med. 2013, 19, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.M.; Curran, T. The Hedgehog’s tale: Developing strategies for targeting cancer. Nat. Rev. Cancer 2011, 11, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Briscoe, J.; Therond, P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013, 14, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Kobune, M.; Takimoto, R.; Murase, K.; Iyama, S.; Sato, T.; Kikuchi, S.; Kawano, Y.; Miyanishi, K.; Sato, Y.; Niitsu, Y.; et al. Drug resistance is dramatically restored by hedgehog inhibitors in CD34+ leukemic cells. Cancer Sci. 2009, 100, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.A.; Copland, M. Targeting hedgehog in hematologic malignancy. Blood 2012, 119, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Mar, B.G.; Amakye, D.; Aifantis, I.; Buonamici, S. The controversial role of the Hedgehog pathway in normal and malignant hematopoiesis. Leukemia 2011, 25, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Dierks, C.; Beigi, R.; Guo, G.R.; Zirlik, K.; Stegert, M.R.; Manley, P.; Trussell, C.; Schmitt-Graeff, A.; Landwerlin, K.; Veelken, H.; et al. Expansion of BCR-ABL-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell 2008, 14, 238–249. [Google Scholar] [CrossRef] [PubMed]
- Krause, D.S.; Van Etten, R.A. Right on target: Eradicating leukemic stem cells. Trends Mol. Med. 2007, 13, 470–481. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y. Overview: Cancer Stem Cell and Tumor Environment. Oncology 2015, 89 (Suppl. S1), 22–24. [Google Scholar] [CrossRef] [PubMed]
- Coombs, C.C.; Tallman, M.S.; Levine, R.L. Molecular therapy for acute myeloid leukaemia. Nat. Rev. Clin. Oncol. 2016, 13, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, G.; Oehler, V.G.; Papayannidis, C.; Courtney, R.; Shaik, M.N.; Zhang, X.; O’Connell, A.; McLachlan, K.R.; Zheng, X.; Radich, J.; et al. Treatment with PF-04449913, an oral smoothened antagonist, in patients with myeloid malignancies: A phase 1 safety and pharmacokinetics study. Lancet Haematol. 2015, 2, e339–e346. [Google Scholar] [CrossRef]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.N.; Borges, I.; Ruiz I Altaba, A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef] [PubMed]
- Satheesha, S.; Manzella, G.; Bovay, A.; Casanova, E.A.; Bode, P.K.; Belle, R.; Feuchtgruber, S.; Jaaks, P.; Dogan, N.; Koscielniak, E.; et al. Targeting hedgehog signaling reduces self-renewal in embryonal rhabdomyosarcoma. Oncogene 2016, 35, 2020–2030. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.M.; Vass, J.K.; Holyoake, T.L.; Graham, G.J. Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells 2007, 25, 3111–3120. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y.; Yamamoto, K.; Kiyoi, H.; Ueda, R.; Saito, H.; Naoe, T. Different antiapoptotic pathways between wild-type and mutated FLT3: Insights into therapeutic targets in leukemia. Blood 2003, 102, 2969–2975. [Google Scholar] [CrossRef] [PubMed]
- Minami, Y.; Stuart, S.A.; Ikawa, T.; Jiang, Y.; Banno, A.; Hunton, I.C.; Young, D.J.; Naoe, T.; Murre, C.; Jamieson, C.H.; et al. BCR-ABL-transformed GMP as myeloid leukemic stem cells. Proc. Natl. Acad. Sci. USA 2008, 105, 17967–17972. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, N.; Minami, Y.; Kakiuchi, S.; Kuwatsuka, Y.; Hayakawa, F.; Jamieson, C.; Kiyoi, H.; Naoe, T. Small-molecule Hedgehog inhibitor attenuates leukemia-initiation potential in acute myeloid leukemia. Cancer Sci. 2016, 107, 1422–1429. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Lauth, M.; Bergstrom, A.; Shimokawa, T.; Toftgard, R. Inhibition of GLImediated transcription and tumor cell growth by small-molecule antagonists. Proc. Natl. Acad. Sci. USA 2007, 104, 8455–8460. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.Z.; Peng, L.F. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol. Biosyst. 2010, 6, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Campbell, V.; Copland, M. Hedgehog signaling in cancer stem cells: A focus on hematological cancers. Stem Cells Cloning 2015, 16, 27–38. [Google Scholar]
- Kim, J.; Tang, J.Y.; Gong, R.; Kim, J.; Lee, J.J.; Clemons, K.V.; Chong, C.R.; Chang, K.S.; Fereshteh, M.; Gardner, D.; et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell 2010, 17, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.J.; Gardner, D.; Beachy, P.A. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc. Natl. Acad. Sci. USA 2010, 107, 13432–13437. [Google Scholar] [CrossRef] [PubMed]
- Li, S.H.; Fu, J.; Watkins, D.N.; Srivastava, R.K.; Shankar, S. Sulforaphane regulates self-renewal of pancreatic cancer stem cells through the modulation of Sonic hedgehog-GLI pathway. Mol. Cell Biochem. 2013, 373, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.N.; Fu, J.; Nall, D.; Rodova, M.; Shankar, S.; Srivastava, R.K. Inhibition of sonic hedgehog pathway and pluripotency maintaining factors regulate human pancreatic cancer stem cell characteristics. Int. J. Cancer 2012, 131, 30–40. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kakiuchi, S.; Minami, Y.; Miyata, Y.; Mizutani, Y.; Goto, H.; Kawamoto, S.; Yakushijin, K.; Kurata, K.; Matsuoka, H.; Minami, H. NANOG Expression as a Responsive Biomarker during Treatment with Hedgehog Signal Inhibitor in Acute Myeloid Leukemia. Int. J. Mol. Sci. 2017, 18, 486. https://doi.org/10.3390/ijms18030486
Kakiuchi S, Minami Y, Miyata Y, Mizutani Y, Goto H, Kawamoto S, Yakushijin K, Kurata K, Matsuoka H, Minami H. NANOG Expression as a Responsive Biomarker during Treatment with Hedgehog Signal Inhibitor in Acute Myeloid Leukemia. International Journal of Molecular Sciences. 2017; 18(3):486. https://doi.org/10.3390/ijms18030486
Chicago/Turabian StyleKakiuchi, Seiji, Yosuke Minami, Yoshiharu Miyata, Yu Mizutani, Hideaki Goto, Shinichiro Kawamoto, Kimikazu Yakushijin, Keiji Kurata, Hiroshi Matsuoka, and Hironobu Minami. 2017. "NANOG Expression as a Responsive Biomarker during Treatment with Hedgehog Signal Inhibitor in Acute Myeloid Leukemia" International Journal of Molecular Sciences 18, no. 3: 486. https://doi.org/10.3390/ijms18030486