Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets
Abstract
:1. Introduction
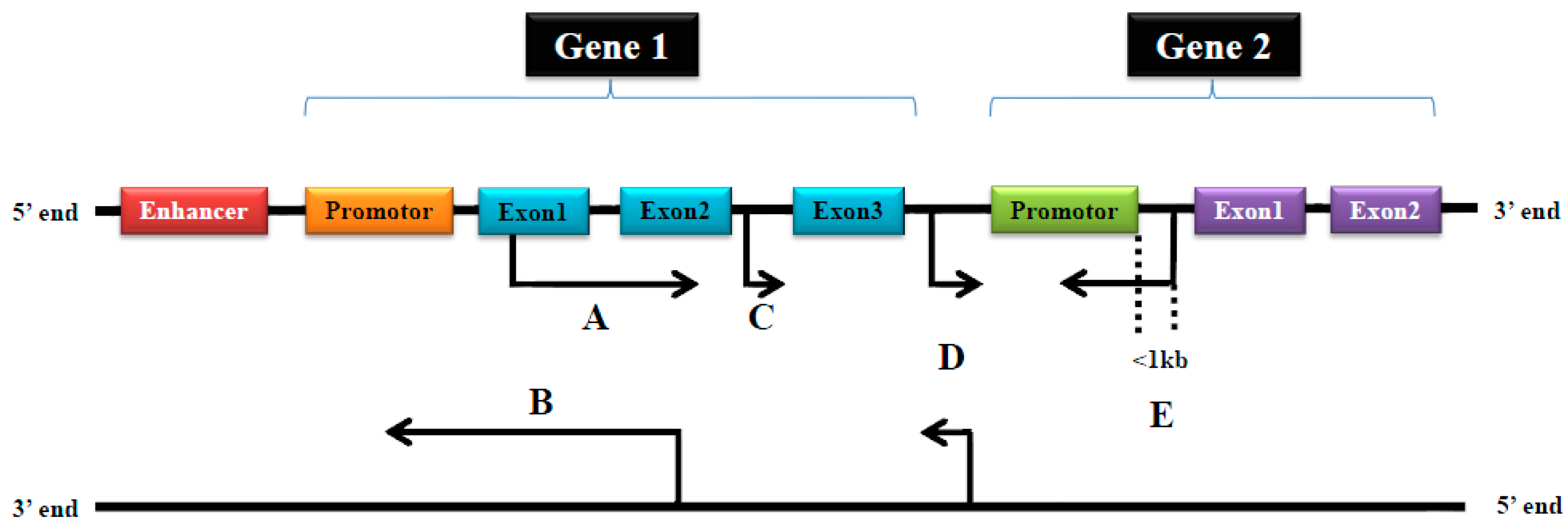
2. Classification and Functions of Long Noncoding RNAs (LncRNAs)
3. The Functions and Mechanisms of LncRNAs in Osteosarcoma
3.1. LncRNAs in Osteosarcoma Cell Proliferation
3.2. LncRNAs in Osteosarcoma Diagnosis
3.3. LncRNAs in Osteosarcoma Metastasis
3.4. LncRNAs in Osteosarcoma Chemoresistance
3.5. LncRNAs in Osteosarcoma Prognosis
4. The Functions and Mechanisms of LncRNAs in Other Sarcomas
4.1. LncRNAs in Chondrosarcoma
4.2. LncRNAs in Ewing’s Sarcoma
4.3. LncRNAs in GISTs
4.4. LncRNAs in Liposarcoma
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Abramowicz, A.; Widlak, P.; Pietrowska, M. Proteomic analysis of exosomal cargo: The challenge of high purity vesicle isolation. Mol. BioSyst. 2016, 12, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Dancsok, A.R.; Asleh-Aburaya, K.; Nielsen, T.O. Advances in sarcoma diagnostics and treatment. Oncotarget 2016, 8, 7068–7093. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Bielack, S.S.; Kempf-Bielack, B.; Branscheid, D.; Carrle, D.; Friedel, G.; Helmke, K.; Kevric, M.; Jundt, G.; Kuhne, T.; Maas, R.; et al. Second and subsequent recurrences of osteosarcoma: Presentation, treatment, and outcomes of 249 consecutive cooperative osteosarcoma study group patients. J. Clin. Oncol. 2009, 27, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Bertone, P.; Stolc, V.; Royce, T.E.; Rozowsky, J.S.; Urban, A.E.; Zhu, X.; Rinn, J.L.; Tongprasit, W.; Samanta, M.; Weissman, S.; et al. Global identification of human transcribed sequences with genome tiling arrays. Science 2004, 306, 2242–2246. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Wang, D.; Qiu, C.; Liu, M.; Chen, X.; Zhang, Q.; Yan, G.; Cui, Q. Lncrnadisease: A database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013, 41, D983–D986. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.F. Non-coding RNAs: Meet thy masters. BioEssays 2010, 32, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wu, D.; Gao, L.; Liu, X.; Jin, Y.; Wang, D.; Wang, T.; Li, X. Competing endogenous RNA networks in human cancer: Hypothesis, validation, and perspectives. Oncotarget 2016, 7, 13479–13490. [Google Scholar] [PubMed]
- Necsulea, A.; Soumillon, M.; Warnefors, M.; Liechti, A.; Daish, T.; Zeller, U.; Baker, J.C.; Grutzner, F.; Kaessmann, H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 2014, 505, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. LincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Yousefi, B. Long non-coding RNAs in cancer drug resistance development. DNA Repair 2016, 45, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.X.; Rastetter, R.H.; Wilhelm, D. Non-coding RNAs: An introduction. Adv. Exp. Med. Biol. 2016, 886, 13–32. [Google Scholar] [PubMed]
- Piatek, M.J.; Werner, A. Endogenous siRNAs: Regulators of internal affairs. Biochem. Soc. Trans. 2014, 42, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.A.; Mitra, A.P.; Triche, T.J. A central role for long non-coding RNA in cancer. Front. Genet. 2012, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Yuan, J.; Li, H.; Li, M.; Zhao, G.; Bu, D.; Zhu, W.; Wu, W.; Chen, R.; Zhao, Y. NONCODEV4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014, 42, D98–D103. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, E.G.; Clark, M.F.; Chen, S.; Cajigas, I.; Leib, D.E.; Kohtz, J.D. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development 2013, 140, 4407–4416. [Google Scholar] [CrossRef] [PubMed]
- Weng, M.; Wu, D.; Yang, C.; Peng, H.; Wang, G.; Wang, T.; Li, X. Noncoding RNAs in the development, diagnosis, and prognosis of colorectal cancer. Transl. Res. 2016, 181, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.M.; Zhou, G.B. Long non-coding RNAs and their roles in non-small-cell lung cancer. Genom. Proteom. Bioinform. 2016, 14, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Cerk, S.; Schwarzenbacher, D.; Adiprasito, J.B.; Stotz, M.; Hutterer, G.C.; Gerger, A.; Ling, H.; Calin, G.A.; Pichler, M. Current status of long non-coding RNAs in human breast cancer. Int. J. Mol. Sci. 2016, 17, E1485. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Yang, Y.; Xu, C.; Xie, Y.; Guo, J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J. Cancer Res. Clin. Oncol. 2016, 142, 2231–2237. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, J.; Chan, M.T.; Wu, W.K. TUG1: A pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016, 49, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, X.; Wu, M.; Lin, C.; Guo, Y.; Tian, B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol. Res. 2016, 23, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cheng, N.; Li, X.; Pan, H.; Li, C.; Ren, S.; Su, C.; Cai, W.; Zhao, C.; Zhang, L.; et al. Correlation of long non-coding RNA H19 expression with cisplatin-resistance and clinical outcome in lung adenocarcinoma. Oncotarget 2017, 8, 2558–2567. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Tang, X.; Li, Y.; Ren, X.; He, Q.; Yang, X.; Zhang, J.; Wang, Y.; Ma, J.; Liu, N. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int. J. Mol. Sci. 2016, 17, 1956. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Li, Y.; Xu, R.; Wang, Y.; Tian, Y.; Chen, W. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: A systematic review and meta-analysis. Oncotarget 2017, 8, 2672–2680. [Google Scholar] [CrossRef] [PubMed]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Feng, Y.; Zhang, D.; Zhao, S.D.; Hu, Z.; Greshock, J.; Zhang, Y.; Yang, L.; Zhong, X.; Wang, L.P.; et al. A functional genomic approach identifies fal1 as an oncogenic long noncoding RNA that associates with bmi1 and represses p21 expression in cancer. Cancer Cell 2014, 26, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Morlando, M.; Ballarino, M.; Fatica, A. Long non-coding RNAs: New players in hematopoiesis and leukemia. Front. Med. 2015, 2, 23. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yan, P.; Lu, J.; Song, G.; Zhu, Y.; Li, Z.; Zhao, Y.; Shen, B.; Huang, X.; Zhu, H.; et al. Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 2015, 16, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, I.A.; Mattick, J.S.; Mehler, M.F. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010, 1338, 20–35. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Houseley, J.; Rubbi, L.; Grunstein, M.; Tollervey, D.; Vogelauer, M. A ncRNA modulates histone modification and mRNA induction in the yeast gal gene cluster. Mol. Cell 2008, 32, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Sissons, H.A. The WHO classification of bone tumors. Recent Results Cancer Res. 1976, 104–108. [Google Scholar]
- Damron, T.A.; Ward, W.G.; Stewart, A. Osteosarcoma, chondrosarcoma, and ewing’s sarcoma: National cancer data base report. Clin. Orthop. Relat. Res. 2007, 459, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.H.; Wang, W.; Yeung, W.; Deng, Y.; Yuan, P.; Mak, K.K. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene 2014, 33, 4857–4866. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Liang, G.; Yuan, B.; Yang, C.; Gao, R.; Zhou, X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3k/Akt pathway. Tumour Biol. 2015, 36, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Zhang, Z. Baicalein suppresses the viability of MG-63 osteosarcoma cells through inhibiting c-MYC expression via Wnt signaling pathway. Mol. Cell. Biochem. 2015, 405, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Geng, P.L.; Yin, P.; Wang, X.L.; Jia, J.P.; Yao, J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac. J. Cancer Prev. 2013, 14, 2311–2315. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.H.; Cao, Y.M.; Huang, Y.; Shi, Q.W.; Guo, J.H.; Fan, Z.W.; Li, J.G.; Chen, B.W.; Wu, B.Y. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging mir-9-5p and regulating POU2f1 expression. Tumour Biol. 2016, 37, 15031–15041. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yao, J.; Meng, H.; Yu, Z.; Wang, Z.; Yuan, X.; Chen, H.; Wang, A. A novel long non-coding RNA, hypoxia-inducible factor-2α promoter upstream transcript, functions as an inhibitor of osteosarcoma stem cells in vitro. Mol. Med. Rep. 2015, 11, 2534–2540. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; He, H.; Xiao, W.; Liu, Q.; Deng, Z.; Lu, Y.; Wang, Q.; Zheng, Q.; Li, Y. MALAT1 promotes osteosarcoma development by targeting TGFA via miR376a. Oncotarget 2016, 7, 54733–54743. [Google Scholar] [PubMed]
- Cai, X.; Liu, Y.; Yang, W.; Xia, Y.; Yang, C.; Yang, S.; Liu, X. Long noncoding RNA MALAT1 as a potential therapeutic target in osteosarcoma. J. Orthop. Res. 2016, 34, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Su, Y.; Yang, Q.; Lv, D.; Zhang, W.; Tang, K.; Wang, H.; Zhang, R.; Liu, Y. Overexpression of long non-coding RNA HOTAIR promotes tumor growth and metastasis in human osteosarcoma. Mol. Cells 2015, 38, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Min, J. Role of long noncoding RNA hotair in the growth and apoptosis of osteosarcoma cell MG-63. BioMed Res. Int. 2016, 2016, 5757641. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cao, L.; Hang, D.; Wang, F.; Wang, Q. Long non-coding RNA HOTTIP is upregulated and associated with poor prognosis in patients with osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 11414–11420. [Google Scholar] [PubMed]
- Min, L.; Hong, S.; Duan, H.; Zhou, Y.; Zhang, W.; Luo, Y.; Shi, R.; Tu, C. Antidifferentiation noncoding RNA regulates the proliferation of osteosarcoma cells. Cancer Biother. Radiopharm. 2016, 31, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yang, C.; Xu, J.; Feng, Y.; Wang, L.; Cui, T. Long noncoding RNA EWSAT1 promotes osteosarcoma cell growth and metastasis through suppression of MEG3 expression. DNA Cell Biol. 2016, 35, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, X.; Fu, C.; Wang, X.; Zou, J.; Hua, H.; Bi, Z. Long noncoding RNA FGFR3-AS1 promotes osteosarcoma growth through regulating its natural antisense transcript FGFR3. Mol. Biol. Rep. 2016, 43, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, F.; Zhao, J.; Li, B.; Liang, Y.; Pan, W.; Zhang, S.; Wang, X.; Zheng, D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget 2016, 7, 82620–82633. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Wang, P.; Feng, S.; Xue, Y.; Li, Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016, 37, 4065–4073. [Google Scholar] [CrossRef] [PubMed]
- Cong, M.; Li, J.; Jing, R.; Li, Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016, 37, 9441–9450. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, P.; Ruan, W.H. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J. Bone Oncol. 2016, 5, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ding, H.; He, E.; Chen, J.; Li, M. Overexpression of long non-coding RNA MFI2 promotes cell proliferation and suppresses apoptosis in human osteosarcoma. Oncol. Rep. 2016, 36, 2033–2040. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Yang, X.; Li, Z.; Jiang, C.; Song, D.; Yan, W.; Liu, T.; Wu, Z.; Kong, J.; Wei, H.; et al. P50-associated COX-2 extragenic RNA (PACER) overexpression promotes proliferation and metastasis of osteosarcoma cells by activating COX-2 gene. Tumour Biol. 2016, 37, 3879–3886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Hou, W.; Tao, J.; Zhao, Y.; Wan, G.; Ma, C.; Xu, H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am. J. Transl. Res. 2016, 8, 3503–3512. [Google Scholar] [PubMed]
- Chen, F.; Mo, J.; Zhang, L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 2016, 37, 13403–13412. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lin, J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am. J. Transl. Res. 2016, 8, 4095–4105. [Google Scholar] [PubMed]
- Marino-Enriquez, A.; Bovee, J.V. Molecular pathogenesis and diagnostic, prognostic and predictive molecular markers in sarcoma. Surg. Pathol. Clin. 2016, 9, 457–473. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Liu, L.H.; Li, J.; Chen, Y.; Jiang, X.W.; Ouyang, Y.R.; Liu, Y.Q.; Zhong, H.; Li, H.; Xiao, T. Microarray expression profile of long noncoding rnas in human osteosarcoma. Biochem. Biophys. Res. Commun. 2013, 433, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, F.; Fei, Z.; Zhao, J.; Liang, Y.; Pan, W.; Liu, X.; Zheng, D. Genetic variants of lncRNA hotair contribute to the risk of osteosarcoma. Oncotarget 2016, 7, 19928–19934. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Li, M.; Zhang, L.; Huang, M.; Lei, J.B.; Fu, G.H.; Liu, C.X.; Lai, Q.W.; Chen, Q.Q.; Wang, Y.L. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosarcoma. Tumour Biol. 2016, 37, 4445–4455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Zhang, G.; Li, S.; Duan, J.; Cheng, J.; Ding, G.; Zhou, C.; Zhang, J.; Luo, P.; et al. Osteosarcoma metastasis: Prospective role of ezrin. Tumour Biol. 2014, 35, 5055–5059. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.H.; Yang, L.B.; Geng, X.L.; Wang, R.; Zhang, Z.C. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 2994–3000. [Google Scholar] [PubMed]
- Wei, X.; Wang, C.; Ma, C.; Sun, W.; Li, H.; Cai, Z. Long noncoding RNA ANRIL is activated by hypoxia-inducible factor-1α and promotes osteosarcoma cell invasion and suppresses cell apoptosis upon hypoxia. Cancer Cell Int. 2016, 16, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.P.; Zhang, C.L.; Shen, G.Q.; Zhu, Z.S. Long noncoding rna expression profiles of the doxorubicin-resistant human osteosarcoma cell line mg63/dxr and its parental cell line MG63 as ascertained by microarray analysis. Int. J. Clin. Exp. Pathol. 2015, 8, 8754–8773. [Google Scholar] [PubMed]
- Zhang, C.L.; Zhu, K.P.; Shen, G.Q.; Zhu, Z.S. A long non-coding RNA contributes to doxorubicin resistance of osteosarcoma. Tumour Biol. 2016, 37, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, L.; Zheng, X.; Zhong, W.; Tian, X.; Yin, B.; Tian, K.; Zhang, W. Long non-coding RNA linc00161 sensitises osteosarcoma cells to cisplatin-induced apoptosis by regulating the mir-645-ifit2 axis. Cancer Lett. 2016, 382, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, L.; Wang, Q. Overexpression of long non-coding RNA HOTTIP increases chemoresistance of osteosarcoma cell by activating the Wnt/β-catenin pathway. Am. J. Transl. Res. 2016, 8, 2385–2393. [Google Scholar] [PubMed]
- Uzan, V.R.; Lengert, A.; Boldrini, E.; Penna, V.; Scapulatempo-Neto, C.; Scrideli, C.A.; Filho, A.P.; Cavalcante, C.E.; de Oliveira, C.Z.; Lopes, L.F.; et al. High expression of hulc is associated with poor prognosis in osteosarcoma patients. PLoS ONE 2016, 11, e0156774. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, X.; Xue, R.; Zhang, Y.; Zhang, X.; Lu, J.; Zhang, Z.; Xue, L. Combined over-expression of the hypoxia-inducible factor 2α gene and its long non-coding RNA predicts unfavorable prognosis of patients with osteosarcoma. Pathol. Res. Pract. 2016, 212, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.T.; Lian, D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3561–3565. [Google Scholar] [PubMed]
- Xia, W.K.; Lin, Q.F.; Shen, D.; Liu, Z.L.; Su, J.; Mao, W.D. Clinical implication of long noncoding RNA 91H expression profile in osteosarcoma patients. OncoTargets Ther. 2016, 9, 4645–4652. [Google Scholar]
- Tian, Z.Z.; Guo, X.J.; Zhao, Y.M.; Fang, Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15138–15142. [Google Scholar] [PubMed]
- Bao, X.; Ren, T.; Huang, Y.; Sun, K.; Wang, S.; Liu, K.; Zheng, B.; Guo, W. Knockdown of long non-coding RNA HOTAIR increases miR-454-3p by targeting Stat3 and Atg12 to inhibit chondrosarcoma growth. Cell Death Dis. 2017, 8, e2605. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Ge, Y.; Guo, L.; Huang, L. Potential approaches to the treatment of ewing’s sarcoma. Oncotarget 2016, 8, 5523–5539. [Google Scholar] [CrossRef] [PubMed]
- Marques Howarth, M.; Simpson, D.; Ngok, S.P.; Nieves, B.; Chen, R.; Siprashvili, Z.; Vaka, D.; Breese, M.R.; Crompton, B.D.; Alexe, G.; et al. Long noncoding RNA ewsat1-mediated gene repression facilitates ewing sarcoma oncogenesis. J. Clin. Investig. 2014, 124, 5275–5290. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.L.; Sanon, M.; Taylor, D.C.; Coombs, J.; Bollu, V.; Sirulnik, L. Epidemiology, survival, and costs of localized gastrointestinal stromal tumors. Int. J. Gen. Med. 2011, 4, 121–130. [Google Scholar] [PubMed]
- Von Mehren, M. Management of gastrointestinal stromal tumors. Surg. Clin. N. Am. 2016, 96, 1059–1075. [Google Scholar] [CrossRef] [PubMed]
- Goswami, D.; Gurule, S.; Lahiry, A.; Anand, A.; Khuroo, A.; Monif, T. Clinical development of imatinib: An anticancer drug. Future Sci. OA 2016, 2, Fso92. [Google Scholar] [CrossRef] [PubMed]
- Ben Ami, E.; Demetri, G.D. A safety evaluation of imatinib mesylate in the treatment of gastrointestinal stromal tumor. Expert Opin. Drug Saf. 2016, 15, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Rutkowski, P.; Hompes, D. Combined therapy of gastrointestinal stromal tumors. Surg. Oncol. Clin. N. Am. 2016, 25, 735–759. [Google Scholar] [CrossRef] [PubMed]
- Niinuma, T.; Suzuki, H.; Nojima, M.; Nosho, K.; Yamamoto, H.; Takamaru, H.; Yamamoto, E.; Maruyama, R.; Nobuoka, T.; Miyazaki, Y.; et al. Upregulation of mir-196a and hotair drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012, 72, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Krikelis, D.; Judson, I. Role of chemotherapy in the management of soft tissue sarcomas. Expert Rev. Anticancer Ther. 2010, 10, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Crago, A.M.; Singer, S. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr. Opin. Oncol. 2011, 23, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhang, Y.; Hou, Y.; Tong, H.; Zhuang, R.; Ji, Z.; Wang, B.; Zhou, Y.; Lu, W. A novel long noncoding RNA PILRLS promote proliferation through TCL1A by activing MDM2 in retroperitoneal liposarcoma. Oncotarget 2017. [Google Scholar] [CrossRef] [PubMed]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Reczko, M.; Maragkakis, M.; Dalamagas, T.M.; Hatzigeorgiou, A.G. Diana-lncbase: Experimentally verified and computationally predicted microrna targets on long non-coding RNAs. Nucleic Acids Res. 2013, 41, D239–D245. [Google Scholar] [CrossRef] [PubMed]


| LncRNA Name | Expression in OS | OS Cells | Potential Mechanism | Tumor Cell Proliferation | Ref. |
|---|---|---|---|---|---|
| TUG1 | High | U2OS cells | Inhibit apoptosis | Promote | [55] |
| U2OS and Saos-2 cells | TUG1/miR-9-5p/ POU2F1 pathway | [56] | |||
| H19 | High | Kios5 cells | Hh signaling pathway and the oncogene Yap1 | Promote | [52] |
| HIF2PUT | Low | SAOS2, MG63, U2OS and OS-732 cells | Control HIF-2α expression | Inhibit | [57] |
| BANCR | Low | MG63 cells | JNK and Wnt/β-catenin signaling pathway | Inhibit | [54] |
| MALAT1 | High | U2OS and Saos2 cells | PI3K/AKT signaling pathway | Promote | [53] |
| Saos2, MG63, U2OS cells | Target TGFA promotion via Inhibiting MIR376A | [58] | |||
| U2OS, HOS, 143B and MG63 cells | Increase RhoA and its downstream effectors ROCKs | [59] | |||
| HOTAIR | High | U2OS, HOS, 143B and MG63 cells | Activate MMP2 and MMP9 | Promote | [60] |
| MG63 cells | Upregulate TGF-β and Bcl-2, upregulate p53 and TNF-α | [61] | |||
| HOTTIP | High | MG-63 and HOS cells | - | Promote | [62] |
| ANCR | High | U2OS and Saos2 cells | Related to p21,CDK2 | Promote | [63] |
| EWSAT1 | High | MG63 and HOS cells | Inhibit MEG3 expression | Promote | [64] |
| FGFR3-AS1 | High | MG63 and U2OS cells | Upregulate FGFR3 expression | Promote | [65] |
| PVT1 | High | KHOS, 143b, LM7, U2OS, and MG63 cells | Increase BCL2 and CCND1 protein expression via negatively regulating miR-195 | Promote | [66] |
| SNHG12 | High | Saos-2, MG63 and U2OS cells | Upregulate AMOT mRNA expression | Promote | [67] |
| TUSC7 | Low | HOS and MG63 | Promote apoptosis | Inhibit | [68] |
| UCA1 | High | HOS, Saos-2, MG63, U2OS cells | Inhibit apoptosis | Promote | [69] |
| MFI2 | High | SAOS-2, MG63, and U2OS cells | Upregulate FOXP4 | Promote | [70] |
| PACER | High | 143B, MG63, Saos-2, U2OS cells | Activate NF-κB-dependent COX-2 | Promote | [71] |
| HNF1A-AS1 | High | HOS, SaOS2, MG63 and U2OS cells | Wnt/β-catenin pathway | Promote | [72] |
| BCAR4 | High | MG63 and U2OS cells | Activate GLI2 pathway | Promote | [73] |
| ZEB1-AS1 | High | HOS, U2OS, MG-63, and Saos-2 cells | Related to ZEB1 lncRNA | Promote | [74] |
| LncRNA Name | Expression in OS | OS Cells | Potential Mechanism | Metastasis | Ref. |
|---|---|---|---|---|---|
| HIF2PUT | Low | SAOS2, MG63, U2OS and OS-732 cells | Control HIF-2α expression | Inhibit | [57] |
| MALAT1 | High | U2OS and SaO2 cells | PI3K/AKT signaling pathway | Promotes | [53] |
| U2OS, HOS, 143B and MG63 cells | Increase RhoA and its downstream effectors ROCKs | [59] | |||
| HOTAIR | High | U2OS, HOS, 143B and MG63 cells | Activate MMP2 and MMP9 | Promotes | [60] |
| HULC | High | MG-63, U2OS and Saos-2 cells | - | Promotes | [81] |
| HOTTIP | High | MG-63 and HOS cells | - | Promotes | [62] |
| ANRIL | High | HOS and U2OS cells | Activated by HIF-1α | Promotes | [82] |
| EWSAT1 | High | MG63 and HOS cells | Inhibit MEG3 expression | Promotes | [64] |
| PVT1 | High | KHOS, 143b, LM7, U2OS, and MG-63 cells | Increase FASN protein expression via negatively regulating miR-195 | Promotes | [66] |
| SNHG12 | High | Saos-2, MG-63 and U2OS cells | Upregulate AMOT mRNA expression | Promotes | [67] |
| UCA1 | High | HOS, Saos-2, MG-63, U2OS cells | - | Promotes | [69] |
| MFI2 | High | SAOS-2, MG63, and U2OS cells | Upregulate FOXP4 | Promotes | [70] |
| PACER | High | 143B, MG63, Saos-2, U2OS cells | Activate NF-κB-dependent COX-2 | Promotes | [71] |
| HNF1A-AS1 | High | HOS, SaOS2, MG63 and U2OS cells | Wnt/β-catenin pathway | Promotes | [72] |
| BCAR4 | High | MG63 and U2OS cells | Activate GLI2 pathway | Promotes | [73] |
| ZEB1-AS1 | High | HOS, U2OS, MG-63, and Saos-2 cells | Related to ZEB1 lncRNA | Promotes | [74] |
| LncRNA Name | Number of OS Sample | Expression Pattern | Prognosis | Ref. |
|---|---|---|---|---|
| HULC | 78 tissue samples | High | Poor | [81] |
| 33 tissue samples | [87] | |||
| MEG3 | 64 tissue samples | Low | Good | [91] |
| HOTTIP | 68 tissue samples | High | Poor | [62] |
| UCA1 | 135 tissue samples | High | Poor | [69] |
| HIF2PUT | 82 tissue samples | High | Poor | [88] |
| BCAR4 | 60 tissue samples | High | Poor | [73] |
| MALAT1 | 162 tissue samples | High | Poor | [89] |
| TUG1 | 76 tissue samples 29 blood samples | High | Poor | [78] |
| FGFR3-AS1 | 62 tissue samples | High | Poor | [65] |
| TUSC7 | 82 tissue samples | Low | Good | [68] |
| HNF1A-AS1 | 43 tissue samples | High | Poor | [72] |
| 91H | 67 tissue samples | High | Poor | [90] |
| ZEB1-AS1 | 50 tissue samples | High | Poor | [74] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Min, L.; Garbutt, C.; Tu, C.; Hornicek, F.; Duan, Z. Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 731. https://doi.org/10.3390/ijms18040731
Min L, Garbutt C, Tu C, Hornicek F, Duan Z. Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets. International Journal of Molecular Sciences. 2017; 18(4):731. https://doi.org/10.3390/ijms18040731
Chicago/Turabian StyleMin, Li, Cassandra Garbutt, Chongqi Tu, Francis Hornicek, and Zhenfeng Duan. 2017. "Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets" International Journal of Molecular Sciences 18, no. 4: 731. https://doi.org/10.3390/ijms18040731
APA StyleMin, L., Garbutt, C., Tu, C., Hornicek, F., & Duan, Z. (2017). Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets. International Journal of Molecular Sciences, 18(4), 731. https://doi.org/10.3390/ijms18040731







