Polymorphisms within Genes Involved in Regulation of the NF-κB Pathway in Patients with Rheumatoid Arthritis
Abstract
:1. Introduction
2. Results
2.1. Response to Treatment
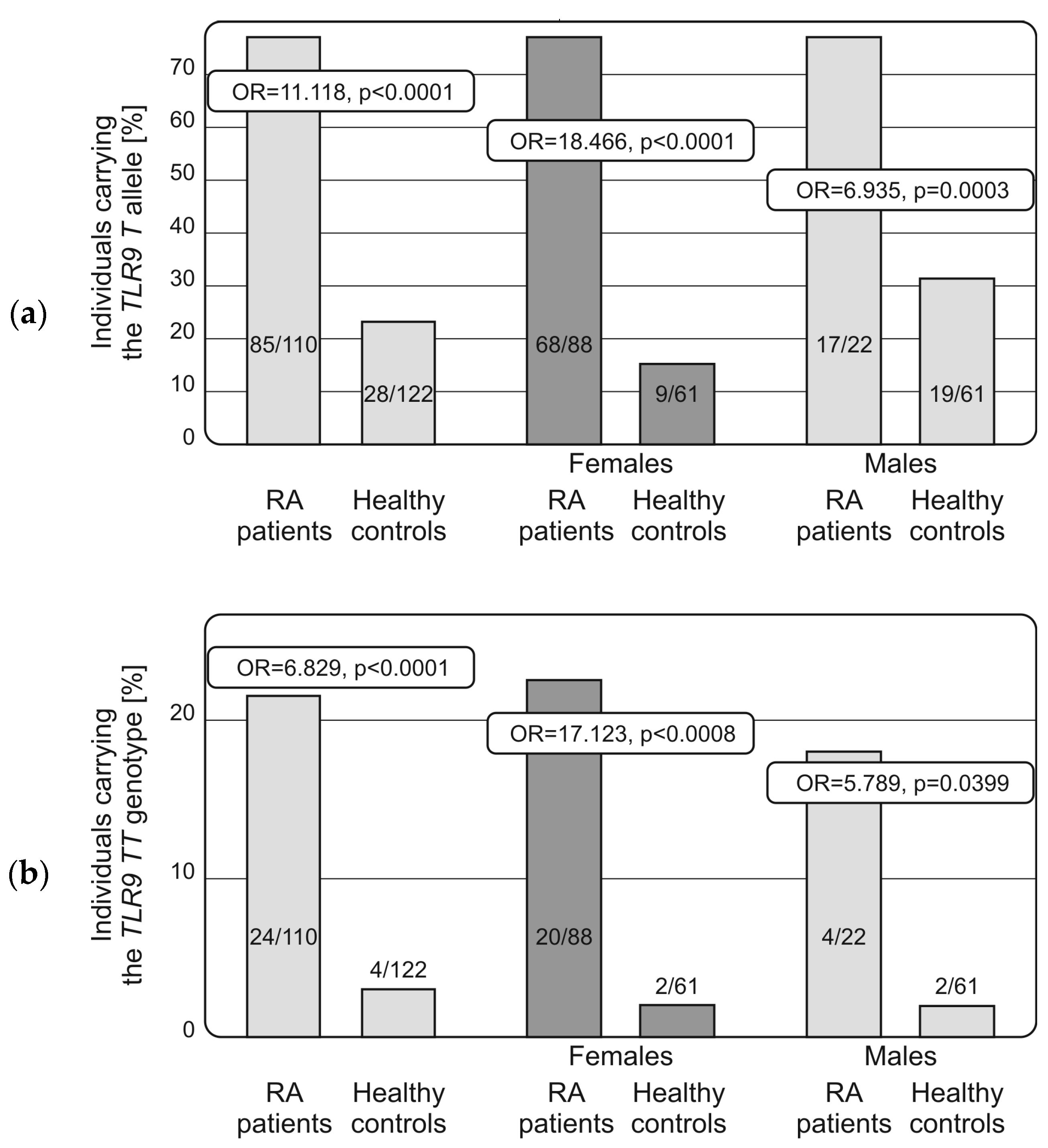
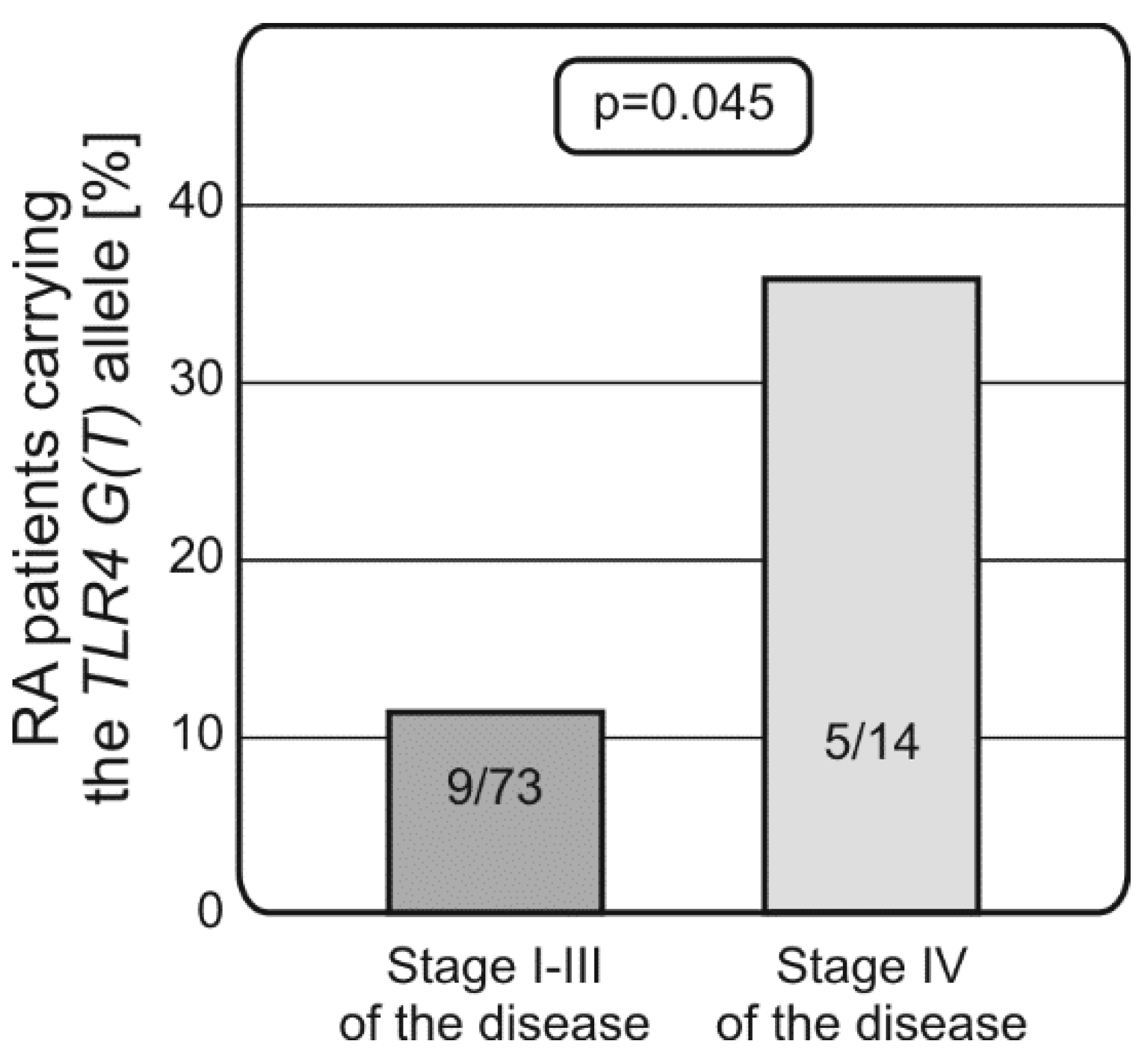
2.2. Distribution of Alleles and Genotypes of TLRs and NF-κB Encoding Genes in RA Patients and Controls, Associations with Disease Susceptibility and Progression
2.3. Genetic Variability within the TLRs and NF-κB1 Genes and Response to Therapy with TNF-α Blocking Agents
3. Discussion
4. Materials and Methods
4.1. Patients and Controls
4.2. DNA Isolation
4.3. NF-κB1 Genotyping
4.4. TLR4 Genotyping
4.5. TLR2 Genotyping
4.6. TLR9 Genotyping
4.7. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DAS 28 | 28-Joint Disease Activity Score |
| DMARD | Disease-Modifying Antirheumatic Drug |
| EULAR | European League Against Rheumatism |
| HWE | Hardy-Weinberg Equilibrium |
| IKK | IκB Kinase |
| MAF | Minor Allele Frequency |
| NF-κB | Nuclear Factor-κB |
| OR | Odds Ratio |
| p | Probability |
| RA | Rheumatoid Arthritis |
| RF | Rheumatoid Factor |
| SNP | Single Nucleotide Polymorphism |
| TLR | Toll-Like Receptor |
| TNF-α | Tumor Necrosis Factor α |
| VAS | Visual Analogue Scale |
References
- Lee, D.M.; Weinblatt, M.E. Rheumatoid arthritis. Lancet 2001, 358, 903–911. [Google Scholar] [CrossRef]
- Shin, H.S.; Jung, Y.S.; Rim, H. Toll-like receptors. Br. J. Med. Med. Res. 2013, 3, 58–68. [Google Scholar] [CrossRef]
- Ospelt, C.; Brentano, F.; Rengel, Y.; Stanczyk, J.; Kolling, C.; Tak, P.P.; Gay, R.E.; Gay, S.; Kyburz, D. Overexpression of toll-like receptors 3 and 4 in synovial tissue from patients with early rheumatoid arthritis: Toll-like receptor expression in early and longstanding arthritis. Arthritis Rheum. 2008, 58, 3684–3692. [Google Scholar] [CrossRef] [PubMed]
- Kuuliala, K.; Orpana, A.; Leirisalo-Repo, M.; Kautiainen, H.; Hurme, M.; Hannonen, P.; Korpela, M.; Möttönen, T.; Paimela, L.; Puolakka, K.; et al. Polymorphism at position +896 of the toll-like receptor 4 gene interferes with rapid response to treatment in rheumatoid arthritis. Ann. Rheum. Dis. 2006, 65, 1241–1243. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef]
- Verstrepen, L.; Bekaert, T.; Chau, T.L.; Tavernier, J.; Chariot, A.; Beyaert, R. TLR-4, IL-1R and TNF-R signaling to NF-κB: Variations on a common theme. Cell. Mol. Life Sci. 2008, 65, 2964–2978. [Google Scholar] [CrossRef] [PubMed]
- Bogunia-Kubik, K.; Świerkot, J.; Malak, A.; Wysoczańska, B.; Nowak, B.; Białowąs, K.; Gębura, K.; Korman, L.; Wiland, P. IL-17A, IL-17F and IL-23R Gene Polymorphisms in Polish Patients with Rheumatoid Arthritis. Arch. Immunol. Ther. Exp. 2015, 63, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Marwa, O.S.; Kalthoum, T.; Wajih, K.; Kamel, H. Association of IL17A and IL17F genes with rheumatoid arthritis disease and the impact of genetic polymorphisms on response to treatment. Immunol. Lett. 2017, 183, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Paradowska-Gorycka, A.; Wojtecka-Lukasik, E.; Trefler, J.; Wojciechowska, B.; Lacki, J.K.; Maslinski, S. Association between IL-17F gene polymorphisms and susceptibility to and severity of rheumatoid arthritis (RA). Scand. J. Immunol. 2010, 72, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Świerkot, J.; Bogunia-Kubik, K.; Nowak, B.; Bialowas, K.; Korman, L.; Gebura, K.; Kolossa, K.; Jeka, S.; Wiland, P. Analysis of associations between polymorphisms within genes coding for tumour necrosis factor (TNF)-α and TNF receptors and responsiveness to TNF-α blockers in patients with rheumatoid arthritis. Jt. Bone Spine 2015, 82, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Emery, P.; Dörner, T. Optimising treatment in rheumatoid arthritis: A review of potential biological markers of response. Ann. Rheum. Dis. 2011, 70, 2063–2070. [Google Scholar] [CrossRef] [PubMed]
- Laska, M.J.; Hansen, B.; Troldborg, A.; Lorenzen, T.; Stengaard-Pedersen, K.; Junker, P.; Nexø, B.A.; Lindegaard, H.M. A non-synonymous single-nucleotide polymorphism in the gene encoding Toll-like Receptor 3 (TLR3) is associated with sero-negative Rheumatoid Arthritis (RA) in a Danish population. BMC Res. Notes 2014, 7, 716. [Google Scholar] [CrossRef] [PubMed]
- Coenen, M.J.; Enevold, C.; Barrera, P.; Schijvenaars, M.M.; Toonen, E.J.; Scheffer, H.; Padyukov, L.; Kastbom, A.; Klareskog, L.; Barton, A.; et al. Genetic variants in toll-like receptors are not associated with rheumatoid arthritis susceptibility or anti-tumour necrosis factor treatment outcome. PLoS ONE 2010, 5, e14326. [Google Scholar] [CrossRef] [PubMed]
- Tizaoui, K.; Naouali, A.; Kaabachi, W.; Hamzaoui, A.; Hamzaoui, K. Association of Toll like receptor Asp299Gly with rheumatoid arthritis risk: A systematic review of case-control studies and meta-analysis. Pathol. Res. Pract. 2015, 211, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Kilding, R.; Akil, M.; Till, S.; Amos, R.; Winfield, J.; Iles, M.M.; Wilson, A.G. A biologically important single nucleotide polymorphism within the toll-like receptor-4 gene is not associated with rheumatoid arthritis. Clin. Exp. Rheumatol. 2003, 21, 340–342. [Google Scholar] [PubMed]
- Lee, Y.H.; Bae, S.C.; Kim, J.H.; Song, G.G. Toll-like receptor polymorphisms and rheumatoid arthritis: A systematic review. Rheumatol. Int. 2014, 34, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.L.; LeVan, T.D.; Yu, F.; Sayles, H.; Sokolove, J.; Robinson, W.; Michaud, K.; Thiele, G.M.; Mikuls, T.R. Associations of toll-like receptor (TLR)-4 single nucleotide polymorphisms and rheumatoid arthritis disease progression: An observational cohort study. Int. Immunopharmacol. 2015, 24, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Devaraju, P.; Gulati, R.; Antony, P.T.; Mithun, C.B.; Negi, V.S. Susceptibility to SLE in South Indian Tamils may be influenced by genetic selection pressure on TLR2 and TLR9 genes. Mol. Immunol. 2015, 64, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.L.; Chen, R.H.; Lin, H.J.; Liu, Y.H.; Chen, W.C.; Tsai, Y.; Wan, L.; Tsai, F.J. Toll-like receptor gene polymorphisms are associated with susceptibility to Graves’ ophthalmopathy in Taiwan males. BMC Med. Genet. 2010, 11, 154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Lv, J.C.; Cheng, W.R.; Yu, L.; Zhao, M.H.; Zhang, H. Association of TLR9 gene polymorphisms with lupus nephritis in a Chinese Han population. Clin. Exp. Rheumatol. 2010, 28, 397–400. [Google Scholar] [PubMed]
- Bharti, D.; Kumar, A.; Mahla, R.S.; Kumar, S.; Ingle, H.; Shankar, H.; Joshi, B.; Raut, A.A.; Kumar, H. The role of TLR9 polymorphism in susceptibility to pulmonary tuberculosis. Immunogenetics 2014, 66, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Wheeless, C.R. Rheumatoid arthritis. In Wheeless’ Textbook of Orthopaedics; Wheeless, C.R., Nunley, J.A., Urbaniak, J.R., Eds.; Data Trace Internet Publishing, LLC: Broklandville, MA, USA, 2012. [Google Scholar]
- Liu, F.; Lu, W.; Qian, Q.; Qi, W.; Hu, J.; Feng, B. Frequency of TLR 2, 4, and 9 gene polymorphisms in Chinese population and their susceptibility to type 2 diabetes and coronary artery disease. J. Biomed. Biotechnol. 2012, 2012, 373945. [Google Scholar] [CrossRef] [PubMed]
| RA Patients (N =) | 87 |
|---|---|
| Sex (female/male); n (%) | 71 (82%)/16 (18%) |
| Age (years) | 50.7 ± 12.3 (range: 17–77) |
| Females (%) | 71 (82%) |
| Disease duration (years) | 12.4 ± 8.3 (range 1–39) |
| Disease onset (years) | 38.8 ± 12.0 (range 15–65) |
| Current smokers (%) | 14 |
| RF+ Rheumatoid factor positive, n (%) | 72 |
| ACPA+/Anti-CCP present, n (%) | 47 |
| Stage, n (%) | |
| 1 | 2 (2.3%) |
| 2 | 20 (23%) |
| 3 | 51 (58.6%) |
| 4 | 14 (16.1%) |
| DAS28 at baseline | 6.59 ± 0.73 (range 5.14–8.05) |
| DAS28 at week 24 | 4.0 ± 1.12 (range 1.97–6.88) |
| anti-TNF drug | |
| etanercept (%) | 44 (50%) |
| adalimumab (%) | 32 (36%) |
| infliximab (%) | 7 (8%) |
| certolizumab pegol (%) | 5 (6%) |
| Glucocorticosteroids % | 79 (mean dose 9.3 mg prednisone daily) |
| Methotrexate % | 71 (mean dose 20.4 mg weekly) |
| Polymorphism | Minor Allele | MAF RA Patients | MAF Controls |
|---|---|---|---|
| NF-κB1 (rs28362491, −94 del/ins ATTG) | del | 0.423 | 0.385 |
| TLR2 (rs111200466, −196/−174 del/ins) | del | 0.168 | 0.179 |
| TLR4 (rs4986790, 13843 A > G) | G | 0.077 | 0.065 |
| TLR4 (rs4986791, 14143 C > T) | T | 0.077 | 0.074 |
| TLR9 (rs5743836; −1237 C > T) | C | 0.135 | 0.083 |
| TLR9 (rs187084; −1486 T > C) | T | 0.495 | 0.131 |
| Polymorphism | RA Patients | Controls | OR, p | ||
|---|---|---|---|---|---|
| n | (%) | n | (%) | ||
| NF-κB1 (rs28362491, −94 del/ins ATTG) | |||||
| del/del | 19 | 17.3 | 14 | 11.1 | |
| del/ins | 55 | 50.0 | 69 | 54.8 | |
| ins/ins | 36 | 32.7 | 43 | 34.1 | |
| del | 74 | 67.3 | 83 | 65.9 | |
| ins | 91 | 82.7 | 112 | 88.9 | |
| HWE | p = 0.798 | p = 0.079 | |||
| TLR2 (rs111200466, −196/−174 del/ins) | |||||
| del/del | 1 | 1.8 | 7 | 5.5 | |
| del/ins | 35 | 31.8 | 31 | 24.6 | |
| ins/ins | 74 | 67.3 | 88 | 69.8 | |
| del | 36 | 32.7 | 39 | 30.9 | |
| ins | 109 | 90.1 | 119 | 94.4 | |
| HWE | p = 0.150 | p = 0.070 | |||
| TLR4 (rs4986790, Asp299Gly, 13843 A > G) | |||||
| AA | 94 | 85.5 | 107 | 87.0 | |
| AG | 15 | 13.6 | 16 | 13.0 | |
| GG | 1 | 0.9 | 0 | 0.0 | |
| A | 109 | 99.1 | 123 | 100.0 | |
| G | 16 | 14.5 | 16 | 13.0 | |
| HWE | p = 0.646 | p = 0.440 | |||
| TLR4 (rs4986791, Thr399Ile, 14,143 C > T) | |||||
| CC | 94 | 85.5 | 104 | 85.2 | |
| CT | 15 | 13.6 | 18 | 14.8 | |
| TT | 1 | 0.9 | 0 | 0.0 | |
| C | 109 | 99.1 | 122 | 100.0 | |
| T | 16 | 14.5 | 0 | 0.0 | |
| HWE | p = 0.646 | p = 0.379 | |||
| TLR9 (rs5743836; −1237 C > T) | |||||
| CC | 1 | 0.9 | 0 | 0.0 | |
| CT | 28 | 25.2 | 21 | 16.7 | |
| TT | 81 | 73.9 | 105 | 83.3 | |
| C | 29 | 26.4 | 21 | 16.7 | (association in females only) # |
| T | 109 | 99.1 | 105 | 83.3 | |
| HWE | p = 0.397 | p = 0.308 | |||
| TLR9 (rs187084; −1486 T > C) | |||||
| TT | 24 | 21.8 | 4 | 3.3 | OR = 6.829, p < 0.0001 |
| TC | 61 | 55.5 | 24 | 19.7 | OR = 4.995, p < 0.0001 |
| CC | 25 | 22.7 | 94 | 77.0 | |
| T | 85 | 77.3 | 28 | 22.9 | OR = 11.118, p < 0.0001 |
| C | 86 | 78.2 | 118 | 96.7 | |
| HWE | p = 0.252 | p = 0.131 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gębura, K.; Świerkot, J.; Wysoczańska, B.; Korman, L.; Nowak, B.; Wiland, P.; Bogunia-Kubik, K. Polymorphisms within Genes Involved in Regulation of the NF-κB Pathway in Patients with Rheumatoid Arthritis. Int. J. Mol. Sci. 2017, 18, 1432. https://doi.org/10.3390/ijms18071432
Gębura K, Świerkot J, Wysoczańska B, Korman L, Nowak B, Wiland P, Bogunia-Kubik K. Polymorphisms within Genes Involved in Regulation of the NF-κB Pathway in Patients with Rheumatoid Arthritis. International Journal of Molecular Sciences. 2017; 18(7):1432. https://doi.org/10.3390/ijms18071432
Chicago/Turabian StyleGębura, Katarzyna, Jerzy Świerkot, Barbara Wysoczańska, Lucyna Korman, Beata Nowak, Piotr Wiland, and Katarzyna Bogunia-Kubik. 2017. "Polymorphisms within Genes Involved in Regulation of the NF-κB Pathway in Patients with Rheumatoid Arthritis" International Journal of Molecular Sciences 18, no. 7: 1432. https://doi.org/10.3390/ijms18071432





