Metabolic Profiling of Dendrobium officinale in Response to Precursors and Methyl Jasmonate
Abstract
:1. Introduction
2. Results
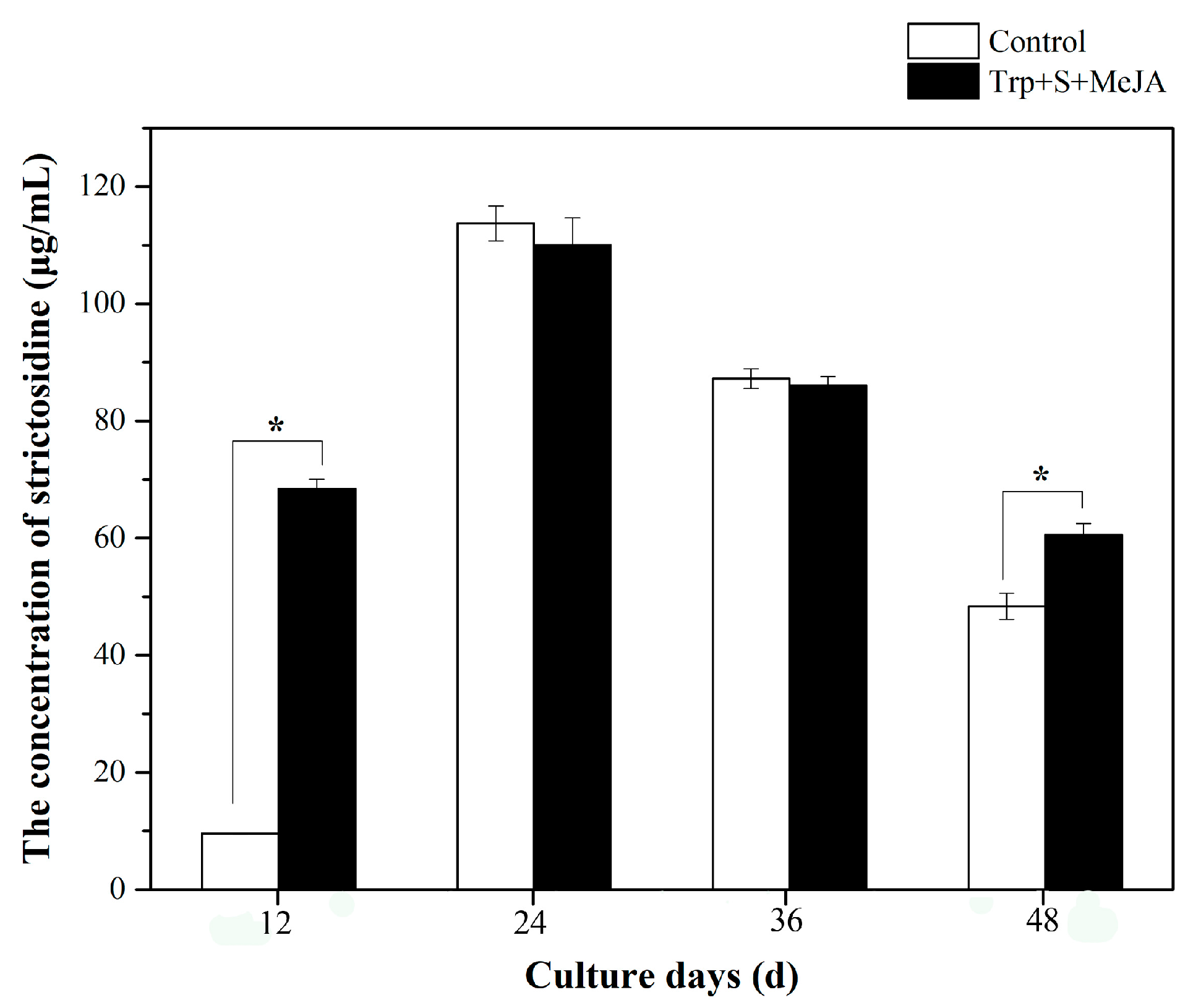
2.1. Effects of MeJA and Its Combination with TIA Precursors on D. officinale PLB Biomass, Alkaloid Content and STR Activity
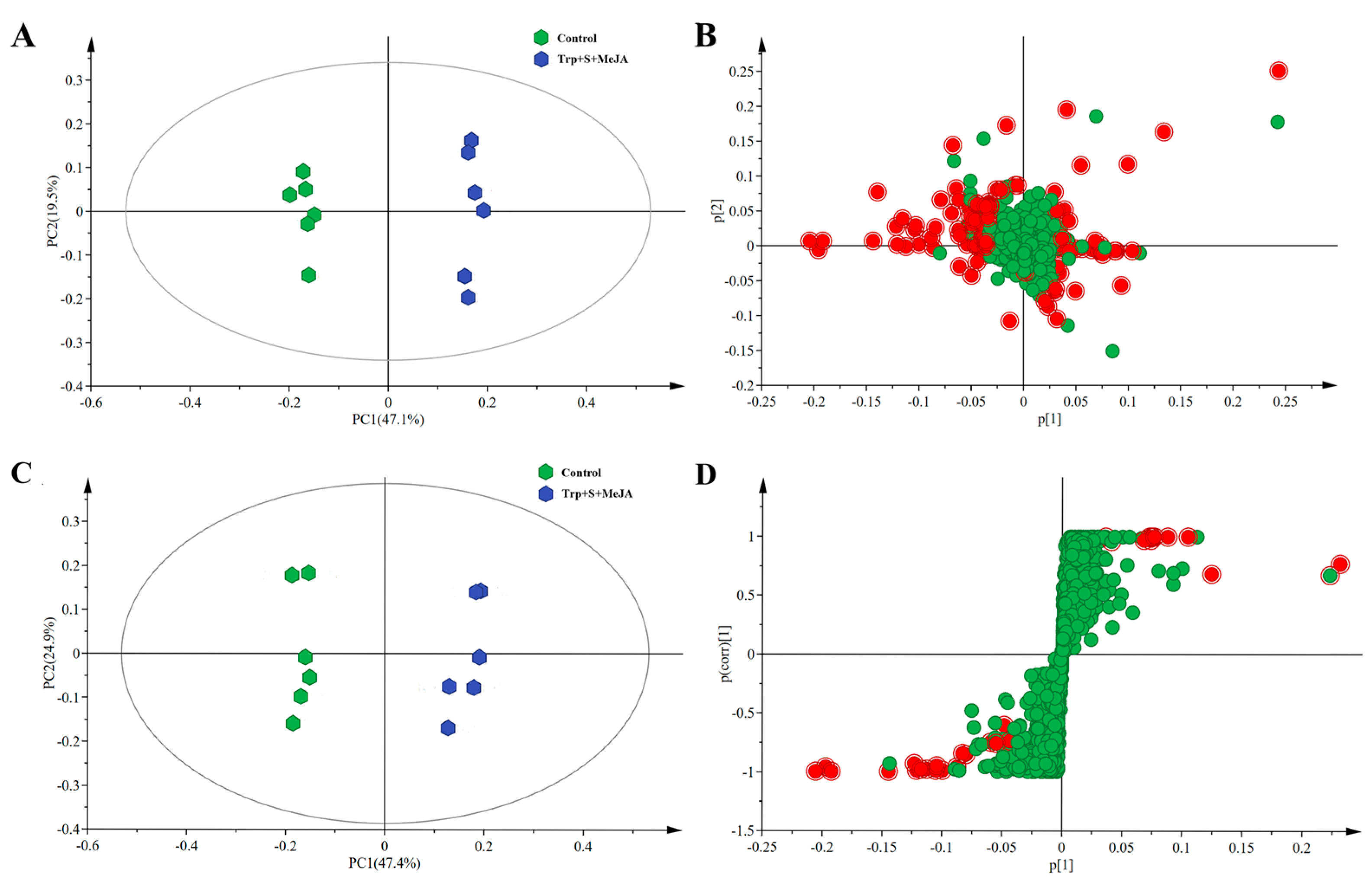
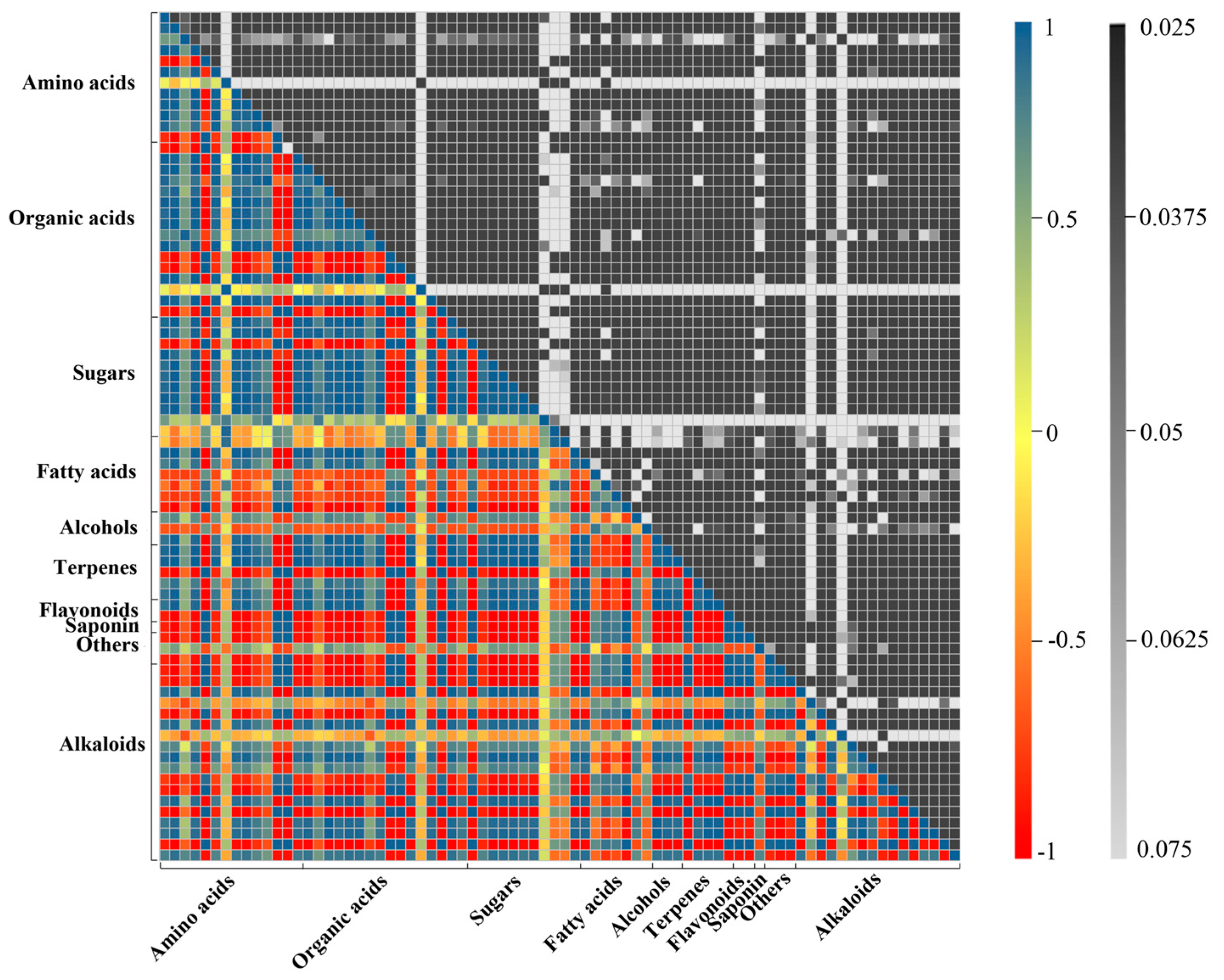
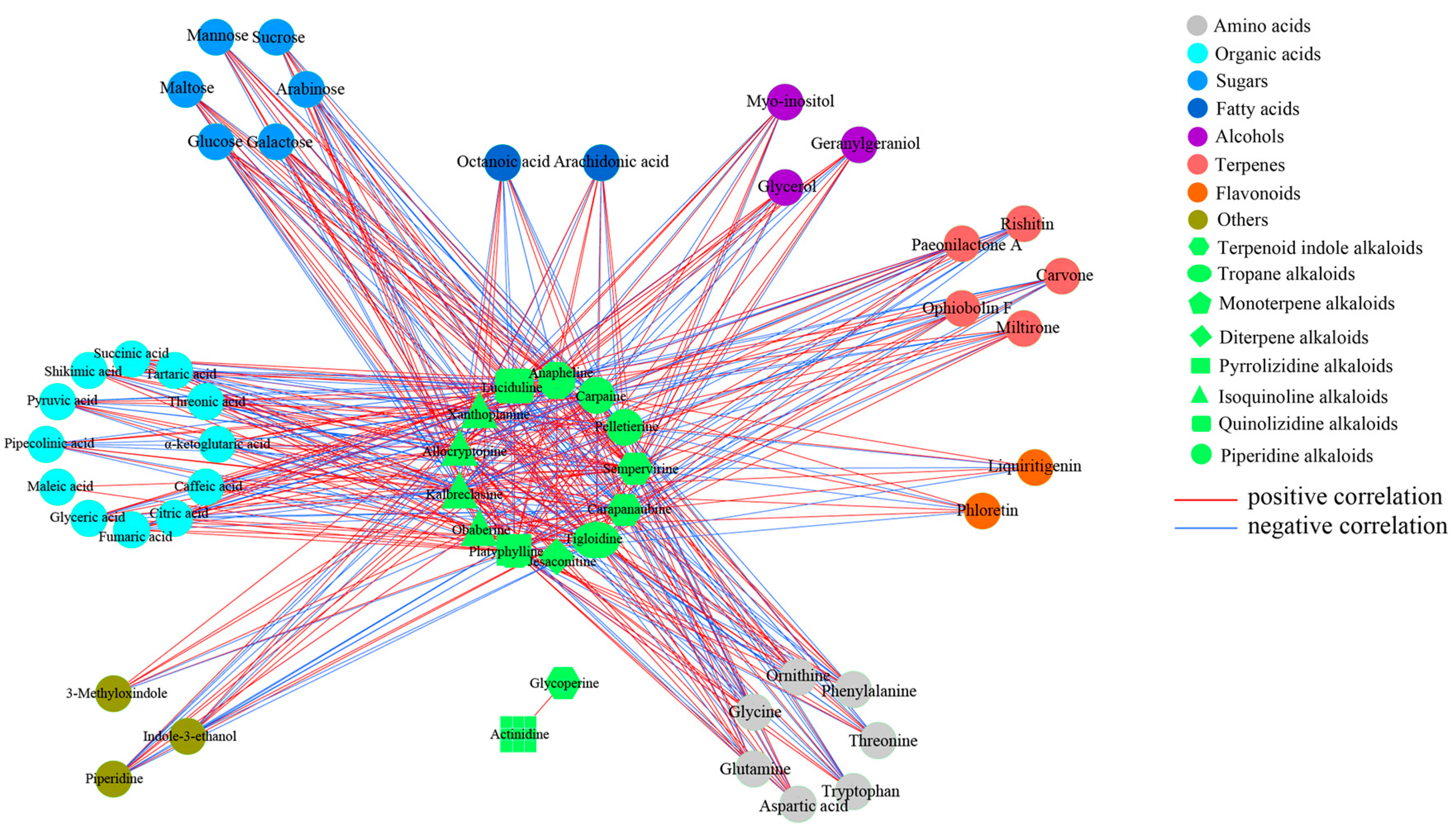
2.2. Metabolic Profiling Analysis of D. officinale PLBs Treated with TIA Precursors and MeJA
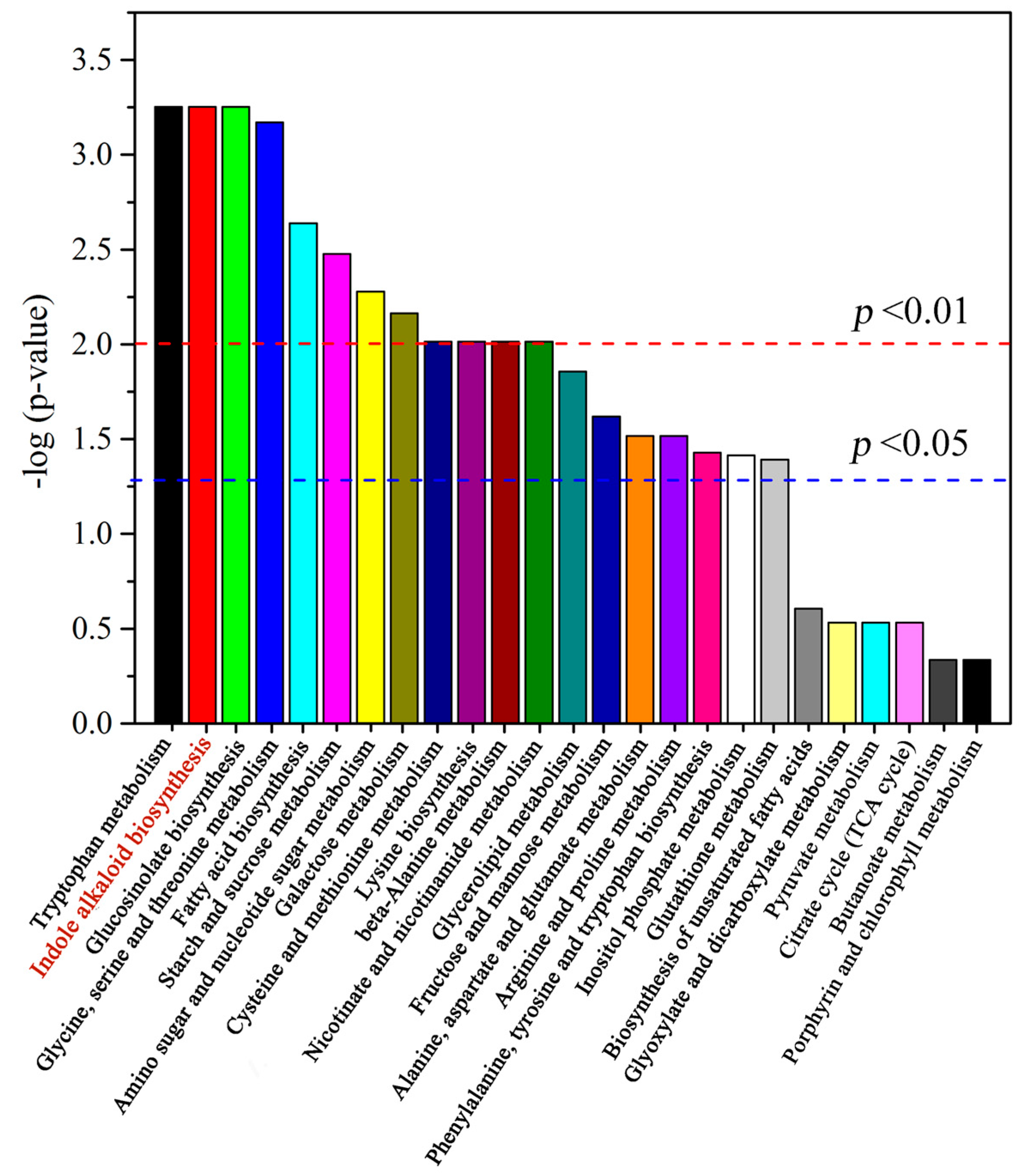
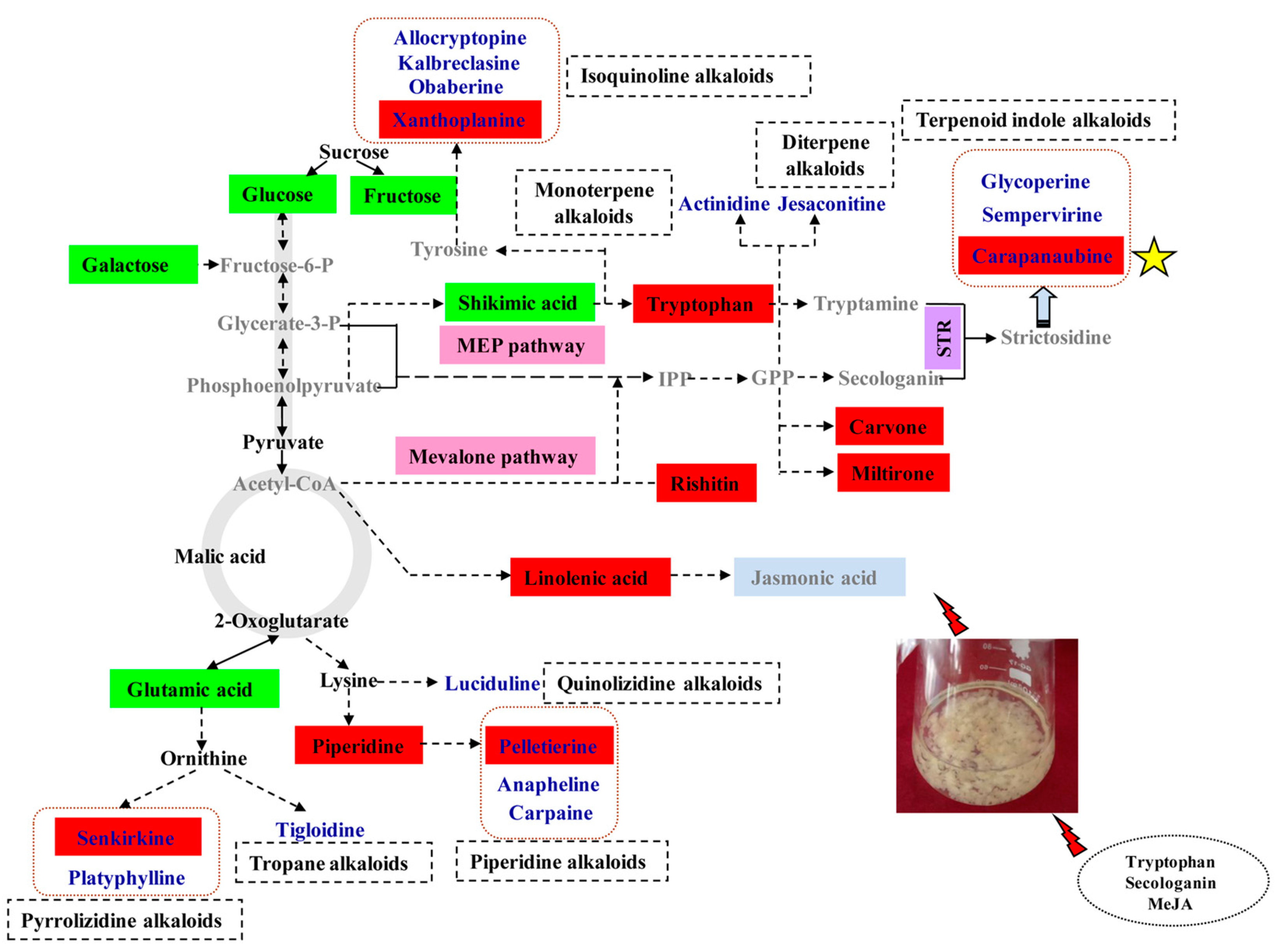
2.3. Alkaloid Biosynthesis Is Affected by TIA Precursors and MeJA Treatment
3. Discussion
3.1. Alkaloid Types and Their Putative Biosynthetic Pathways in D. officinale
3.2. Metabolic Changes in Alkaloid Biosynthesis in D. officinale in Response to TIA Precursors and MeJA Treatment
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Chemicals and Reagents
4.3. Elicitation and Precursor Feeding Experiments
4.4. Determination of Biomass and Total Alkaloid Content
4.5. Strictosidine Synthase Assay
4.6. Metabolite Extraction for GC-MS Analysis
4.7. Metabolite Extraction for LC-MS Analysis
4.8. Instrument Parameters for GC-MS
4.9. Instrument Parameters for LC-MS
4.10. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cui, H.Y.; Murthy, H.N.; Moh, S.H.; Cui, Y.Y.; Paek, K.Y. Establishment of protocorm suspension cultures of Dendrobium candidum for the production of bioactive compounds. Hortic. Environ. Biotechnol. 2015, 1, 114–122. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Sze, S.C.W.; Tong, Y.; Zhang, K.Y. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, C.; Wu, K.; Teixeira da Silva, J.A.; Zeng, S.; Zhang, X.; Yu, Z.; Xia, H.; Duan, J. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 2016, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Zhao, T.; Sheng, Y.; Zheng, T.; Fu, L.; Zhang, Y. Dendrobium officinale Kimura et Migo: A Review on Its Ethnopharmacology, Phytochemistry, Pharmacology, and Industrialization. Evid.-Based Complement. Altern. Med. 2017, 2017, 7436259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, L.; Liu, J.; Liang, J.; Si, J.; Wu, S. Dendrobium officinale, leaves as a new antioxidant source. J. Funct. Foods 2017, 37, 400–415. [Google Scholar] [CrossRef]
- Chen, X.M.; Xiao, S.Y.; Guo, S.X. Comparison of chemical compositions between Dendrobium candidum and Dendrobium nobile. Acta Acad. Med. Sin. 2006, 28, 524–529. [Google Scholar]
- Guo, X.; Li, Y.; Li, C.; Luo, H.; Wang, L.; Qian, J.; Luo, X.; Xiang, L.; Song, J.; Sun, C.; et al. Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 2013, 527, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Wang, X.; Liu, H.; Tian, Y.; Lian, J.; Yang, R.; Hao, S.; Wang, X.; Yang, S.; Li, Q.; et al. The genome of Dendrobium officinale illuminates the biology of the important traditional Chinese orchid herb. Mol. Plant 2015, 8, 922–934. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Guo, H.; Chen, H.; Shi, Y.; Meng, Y.; Lu, J.; Feng, S.; Wang, H. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 2017, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zeng, X.; Sun, C.; Chen, S. Biosynthetic pathway of terpenoid indole alkaloids in Catharanthus roseus. Front. Med. 2014, 8, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Rischer, H.; Oresic, M.; Seppanen-Laakso, T.; Katajamaa, M.; Lammertyn, F.; Ardiles-Diaz, W.; Van Montagu, M.C.; Inze, D.; Oksman-Caldentey, K.M.; Goossens, A. Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc. Natl. Acad. Sci. USA 2006, 103, 5614–5619. [Google Scholar] [CrossRef] [PubMed]
- Skrzypczak-Pietraszek, E.; Słota, J.; Pietraszek, J. The influence of L-phenylalanine, methyl jasmonate and sucrose concentration on the accumulation of phenolic acids in Exacum affine Balf. f. ex Regel shoot culture. Acta Biochim. Pol. 2014, 61, 47–53. [Google Scholar] [PubMed]
- Pauwels, L.; Morreel, K.; De Witte, E.; Lammertyn, F.; Van Montagu, M.; Boerjan, W.; Inze, D.; Goossens, A. Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 2008, 105, 1380–1385. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Jin, M.Y.; Paek, K.Y.; Piao, X.C.; Lian, M.L. An efficient strategy for enhancement of bioactive compounds by protocorm-like body culture of Dendrobium candidum. Ind. Crops Prod. 2016, 84, 121–130. [Google Scholar] [CrossRef]
- Yuan, Z.Q.; Zhang, J.Y.; Liu, T. Enhancement of polysaccharides accumulation in Dendrobium officinale by exogenously applied methyl jasmonate. Biol. Plant. 2017, 61, 438–444. [Google Scholar] [CrossRef]
- Panwar, G.S.; Guru, S.K. Stimulation of reserpine production in the whole plant culture of Rauwolfia serpentina L. by elicitors and precursor feeding. J. Plant Biochem. Biotechnol. 2015, 24, 49–55. [Google Scholar] [CrossRef]
- Whitmer, S.; van der Heijden, R.; Verpoorte, R. Effect of precursor feeding on alkaloid accumulation by a tryptophan decarboxylase over-expressing transgenic cell line T22 of Catharanthus roseus. J. Biotechnol. 2002, 96, 193–203. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Wang, Y.; Abozeid, A.; Zu, Y.G.; Tang, Z.H. The integration of GC-MS and LC-MS to assay the metabolomics profiling in Panax ginseng and Panax quinquefolius reveals a tissue-and species-specific connectivity of primary metabolites and ginsenosides accumulation. J. Pharm. Biomed. Anal. 2017, 135, 176–185. [Google Scholar] [CrossRef] [PubMed]
- Tugizimana, F.; Ncube, E.N.; Steenkamp, P.A.; Dubery, I.A. Metabolomics-derived insights into the manipulation of terpenoid synthesis in Centella asiatica cells by methyl jasmonate. Plant Biotechnol. Rep. 2015, 9, 125–136. [Google Scholar] [CrossRef]
- Chen, Q.; Lu, X.; Guo, X.; Guo, Q.; Li, D. Metabolomics Characterization of Two Apocynaceae Plants, Catharanthus roseus and Vinca minor, Using GC-MS and LC-MS Methods in Combination. Molecules 2017, 22, 997. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Jiao, C.; Sun, S.; Song, C.; Cai, Y.; Lin, Y.; Fan, H.; Zhu, Y. Metabolic analysis of medicinal Dendrobium officinale and Dendrobium huoshanense during different growth years. PLoS ONE 2016, 11, e0146607. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.D.; You, T.; Li, J.; Bai, L.T.; Hao, J.W.; Xu, X.Y. A comparative study of three tissue-cultured Dendrobium species and their wild correspondences by headspace gas chromatography-mass spectrometry combined with chemometric methods. J. Food Drug Anal. 2016, 24, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.H.; Avula, B.; Abe, N.; Wei, F.; Wang, M.; Ma, S.C.; Ali, Z.; Elsohly, M.A.; Khan, I.A. Tandem mass spectrometry for structural identification of sesquiterpene alkaloids from the stems of Dendrobium nobile using LC-QToF. Planta Med. 2016, 82, 622–670. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ding, G.; Li, B.; Guo, S.X. Transcriptome Analysis of Genes Involved in Dendrobine Biosynthesis in Dendrobium nobile Lindl. Infected with Mycorrhizal Fungus MF23 (Mycena sp.). Sci. Rep. 2017, 7, 361. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Wu, Q.; Wang, X.; Wu, L.; Cai, Y.; Lin, Y. Molecular cloning and expression of 1-deoxy-d-xylulose-5-phosphate synthase and 1-deoxy-d-xylulose-5-phosphate reductoisomerase in Dendrobium officinale. Plant Cell Tissue Organ Cult. 2016, 125, 381–385. [Google Scholar] [CrossRef]
- Xu, Y.Y.; Wang, L.; Huang, B.; Xie, X.; Wu, Q.; Shi, J. Comparison of contents of polysaccharides and alkaloids in Dendrobium from different harvest time. West China J. Pharm. Sci. 2014, 29, 288–291. [Google Scholar]
- Kierkegaard, P.; Pilotti, A.M. Studies on orchidaceae alkaloids. XX. The constitution and relative configuration of crepidine, an alkaloid from Dendrobium crepidatum Lindl. Acta Chem. Scand. 1970, 24, 3735–3759. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2011, 40, D109–D114. [Google Scholar] [CrossRef] [PubMed]
- Langel, D.; Ober, D.; Pelser, P.B. The evolution of pyrrolizidine alkaloid biosynthesis and diversity in the Senecioneae. Phytochem. Rev. 2011, 10, 3–74. [Google Scholar] [CrossRef]
- Yamada, Y.; Kokabu, Y.; Chaki, K.; Yoshimoto, T.; Ohgaki, M.; Yoshida, S.; Kato, N.; Koyama, T.; Sato, F. Isoquinoline alkaloid biosynthesis is regulated by a unique bHLH-type transcription factor in Coptis japonica. Plant Cell Physiol. 2011, 52, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Liu, X.; Pan, G.; Hou, X.; Zhang, H.; Yuan, Y. In vitro characterization of a (E)-β-farnesene synthase from Matricaria recutita L. and its up-regulation by methyl jasmonate. Gene 2015, 571, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Memelink, J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016, 34, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Wungsintaweekul, J.; Choo-malee, J.; Charoonratana, T.; Keawpradub, N. Methyl jasmonate and yeast extract stimulate mitragynine production in Mitragyna speciosa (Roxb.) Korth. shoot culture. Biotechnol. Lett. 2012, 34, 1945–1950. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; Singh, S.K.; Patra, B.; Sui, X.; Pattanaik, S.; Yuan, L. A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 2017, 213, 1107–1123. [Google Scholar] [CrossRef] [PubMed]
- Flores-Sanchez, I.J.; Paniagua-Vega, D.; Vera-Reyes, I.; Cerda-García-Rojas, C.M.; Ramos-Valdivia, A.C. Alkaloid biosynthesis and metabolic profiling responses to jasmonic acid elicitation in Hamelia patens plants by NMR-based metabolomics. Metabolomics 2016, 12, 66. [Google Scholar] [CrossRef]
- Wu, J.; Wang, L.; Baldwin, I.T. Methyl jasmonate-elicited herbivore resistance: Does MeJA function as a signal without being hydrolyzed to JA? Planta 2008, 227, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Balusamy, S.R.D.; Rahimi, S.; Sukweenadhi, J.; Kim, Y.J.; Yang, D.C. Exogenous methyl jasmonate prevents necrosis caused by mechanical wounding and increases terpenoid biosynthesis in Panax ginseng. Plant Cell Tissue Organ Cult. 2015, 123, 341–348. [Google Scholar] [CrossRef]
- Miersch, O.; Wasternack, C. Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: Endogenous jasmonates do not induce jasmonate biosynthesis. Biol. Chem. 2000, 381, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Carrari, F.; Baxter, C.; Usadel, B.; Urbanczyk-Wochniak, E.; Zanor, M.I.; Nunes-Nesi, A.; Nikiforova, V.; Centero, D.; Ratzka, A.; Pauly, M.; et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 2006, 142, 1380–1396. [Google Scholar] [CrossRef] [PubMed]
- Hedhili, S.; Courdavault, V.; Giglioli-Guivarc’h, N.; Gantet, P. Regulation of the terpene moiety biosynthesis of Catharanthus roseus terpene indole alkaloids. Phytochem. Rev. 2007, 6, 341–351. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Lundborg, L.; Nordlander, G.; Björklund, N.; Nordenhem, H.; Borg-Karlson, A.K. Methyl Jasmonate-Induced Monoterpenes in Scots Pine and Norway Spruce Tissues Affect Pine Weevil Orientation. J. Chem. Ecol. 2016, 42, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Zhu, S.; Zhao, T.; Zhang, M.; Liu, W.; Chen, M.; Lan, X.; Liao, Z. Enhancement of artemisinin content and relative expression of genes of artemisinin biosynthesis in Artemisia annua by exogenous MeJA treatment. Plant Growth Regul. 2015, 75, 435–441. [Google Scholar] [CrossRef]
- Vaccaro, M.C.; Mariaevelina, A.; Malafronte, N.; De Tommasi, N.; Leone, A. Increasing the synthesis of bioactive abietane diterpenes in Salvia sclarea hairy roots by elicited transcriptional reprogramming. Plant Cell Rep. 2017, 36, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Aleong, J.; Howard, D. Extensions of the duncan’s multiple range test for unbalanced data. J. Appl. Stat. 1985, 12, 83–90. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Di Fiore, S.; Fischer, R. Expression of strictosidine synthase in selected subcellular compartments of tobacco plant. Acta Bot. Sin. 2002, 44, 579–582. [Google Scholar]
- Vera-Reyes, I.; Huerta-Heredia, A.A.; Ponce-Noyola, T.; Flores-Sanchez, I.J.; Esparza-García, F.; Cerda-García-Rojas, C.M.; Trejo-Tapia, G.; Ramos-Valdivia, A.C. Strictosidine-related enzymes involved in the alkaloid biosynthesis of Uncaria tomentosa root cultures grown under oxidative stress. Biotechnol. Prog. 2013, 29, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Akhgari, A.; Yrjönen, T.; Laakso, I.; Vuorela, H.; Oksman-Caldentey, K.M.; Rischer, H. Establishment of transgenic Rhazya stricta hairy roots to modulate terpenoid indole alkaloid production. Plant Cell Rep. 2015, 34, 1939–1952. [Google Scholar] [CrossRef] [PubMed]
- Vinaixa, M.; Schymanski, E.L.; Neumann, S.; Navarro, M.; Salek, R.M.; Yanes, O. Mass spectral databases for LC/MS-and GC/MS-based metabolomics: State of the field and future prospects. TrAC Trends Anal. Chem. 2016, 78, 23–35. [Google Scholar] [CrossRef]
- Meyer, M.R.; Peters, F.T.; Maurer, H.H. Automated mass spectral deconvolution and identification system for GC-MS screening for drugs, poisons, and metabolites in urine. Clin. Chem. 2010, 56, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.; Willett, P.; Klaffke, W.; van Noort, P. Evaluation of similarity measures for searching the dictionary of natural products database. J. Chem. Inf. Comput. Sci. 2003, 43, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Little, J.L.; Williams, A.J.; Pshenichnov, A.; Tkachenko, V. Identification of “known unknowns” utilizing accurate mass data and ChemSpider. J. Am. Soc. Mass Spectrom. 2012, 23, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]



| No. | [A] Tryptophan (µM) | [B] Secologanin (µM) | [C] MeJA (µM) | Total Alkaloid Content (μg/g DW) |
|---|---|---|---|---|
| 1 | 1 (3) | 1 (3) | 1 (100) | 188 |
| 2 | 1 | 2 (6) | 2 (200) | 135 |
| 3 | 1 | 3 (9) | 3 (300) | 190 |
| 4 | 2 (6) | 1 | 2 | 190 |
| 5 | 2 | 2 | 3 | 190 |
| 6 | 2 | 3 | 1 | 257 |
| 7 | 3 (9) | 1 | 3 | 124 |
| 8 | 3 | 2 | 1 | 472 |
| 9 | 3 | 3 | 2 | 177 |
| K1 | 171.000 | 167.667 | 305.667 | |
| K2 | 212.333 | 265.667 | 167.333 | |
| K3 | 258.000 | 208.000 | 168.333 | |
| Range | 87.000 | 98.000 | 138.334 | |
| Order | C > B > A | |||
| Optimal level | A3 | B2 | C1 | |
| Optimal group | A3B2C1 |
| No. | Metabolites | Catogery | VIP | Change Fold (Trp + S + MeJA/Control) | p-Value |
|---|---|---|---|---|---|
| 1 | Tryptophan | Amino acid | 4.57 | 20.65 | ** 7.43 × 10−15 |
| 2 | Glycine | Amino acid | 3.82 | 20.65 | ** 7.43 × 10−15 |
| 3 | Aspartic acid | Amino acid | 4.36 | 0.59 | ** 1.79 × 10−9 |
| 4 | Glutamic acid | Amino acid | 2.18 | 0.54 | * 0.03 |
| 5 | Fructose | Sugar | 6.29 | 0.31 | ** 5.15 × 10−12 |
| 6 | Trehalose | Sugar | 1.51 | 3.03 | ** 5.98 × 10−13 |
| 7 | Glucose | Sugar | 8.97 | 0.51 | ** 4.23 × 10−10 |
| 8 | Mannose | Sugar | 4.92 | 0.72 | ** 9.22 × 10−8 |
| 9 | Galactose | Sugar | 8.41 | 0.47 | ** 9.78 × 10−11 |
| 10 | Arabinose | Sugar | 3.27 | 2.69 | ** 1.43 × 10−12 |
| 11 | Stearic acid | Fatty acid | 11.53 | 1.13 | * 0.02 |
| 12 | Arachidonic acid | Fatty acid | 5.14 | 0.07 | ** 1.29 × 10−7 |
| 13 | Eicosanoic acid | Fatty acid | 6.06 | 1.47 | ** 1.00 × 10−7 |
| 14 | Linolenic acid | Fatty acid | 10.68 | 1.45 | ** 0.003 |
| 15 | Pentadecanoic acid | Fatty acid | 2.45 | 0.83 | ** 0.005 |
| 16 | Glycerol | Alcohol | 5.27 | 0.65 | ** 5.61 × 10−9 |
| 17 | Myo-inositol | Alcohol | 3.79 | 0.77 | ** 7.17 × 10−7 |
| 18 | Malic acid | Organic acid | 2.64 | 0.46 | ** 0.005 |
| 19 | Shikimic acid | Organic acid | 8.61 | 0.78 | ** 9.78 × 10−11 |
| 20 | Carapanaubine | Terpenoid indole alkaloid | 2.98 | 1928.46 | ** 1.77 × 10−7 |
| 21 | Pelletierine | Piperidine alkaloid | 5.38 | 0.57 | ** 3.61 × 10−5 |
| 22 | Senkirkine | Pyrrolizidine alkaloid | 3.69 | 0.68 | ** 0.001 |
| 23 | Xanthoplanine | Isoquinoline alkaloid | 3.34 | 1.13 | * 0.018 |
| 24 | Carvone | Terpene | 3.17 | 8.81 | ** 3.12 × 10−12 |
| 25 | Rishitin | Terpene | 2.45 | 26.74 | ** 4.88 × 10−13 |
| 26 | Miltirone | Terpene | 4.53 | 0.41 | ** 4.25 × 10−6 |
| 27 | Indole-3-ethanol | Other | 3.22 | 2.18 | ** 1.00 × 10−7 |
| 28 | Piperidine | Other | 1.61 | 7.24 | ** 2.37 × 10−14 |
| 29 | Camelliagenin C | Saponin | 3.23 | 0.58 | * 0.013 |
| Level | Factors | ||
|---|---|---|---|
| [A] Tryptophan (µM) | [B] Secologanin (µM) | [C] MeJA (µM) | |
| 1 | 3 | 3 | 100 |
| 2 | 6 | 6 | 200 |
| 3 | 9 | 9 | 300 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, C.; Song, C.; Zheng, S.; Zhu, Y.; Jin, Q.; Cai, Y.; Lin, Y. Metabolic Profiling of Dendrobium officinale in Response to Precursors and Methyl Jasmonate. Int. J. Mol. Sci. 2018, 19, 728. https://doi.org/10.3390/ijms19030728
Jiao C, Song C, Zheng S, Zhu Y, Jin Q, Cai Y, Lin Y. Metabolic Profiling of Dendrobium officinale in Response to Precursors and Methyl Jasmonate. International Journal of Molecular Sciences. 2018; 19(3):728. https://doi.org/10.3390/ijms19030728
Chicago/Turabian StyleJiao, Chunyan, Cheng Song, Siyan Zheng, Yingpeng Zhu, Qing Jin, Yongping Cai, and Yi Lin. 2018. "Metabolic Profiling of Dendrobium officinale in Response to Precursors and Methyl Jasmonate" International Journal of Molecular Sciences 19, no. 3: 728. https://doi.org/10.3390/ijms19030728
APA StyleJiao, C., Song, C., Zheng, S., Zhu, Y., Jin, Q., Cai, Y., & Lin, Y. (2018). Metabolic Profiling of Dendrobium officinale in Response to Precursors and Methyl Jasmonate. International Journal of Molecular Sciences, 19(3), 728. https://doi.org/10.3390/ijms19030728