Selection and Characterization of a DNA Aptamer Specifically Targeting Human HECT Ubiquitin Ligase WWP1
Abstract
:1. Introduction
2. Results
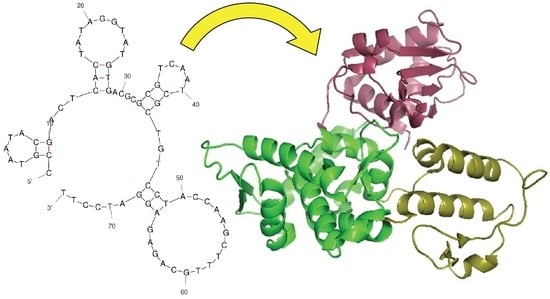
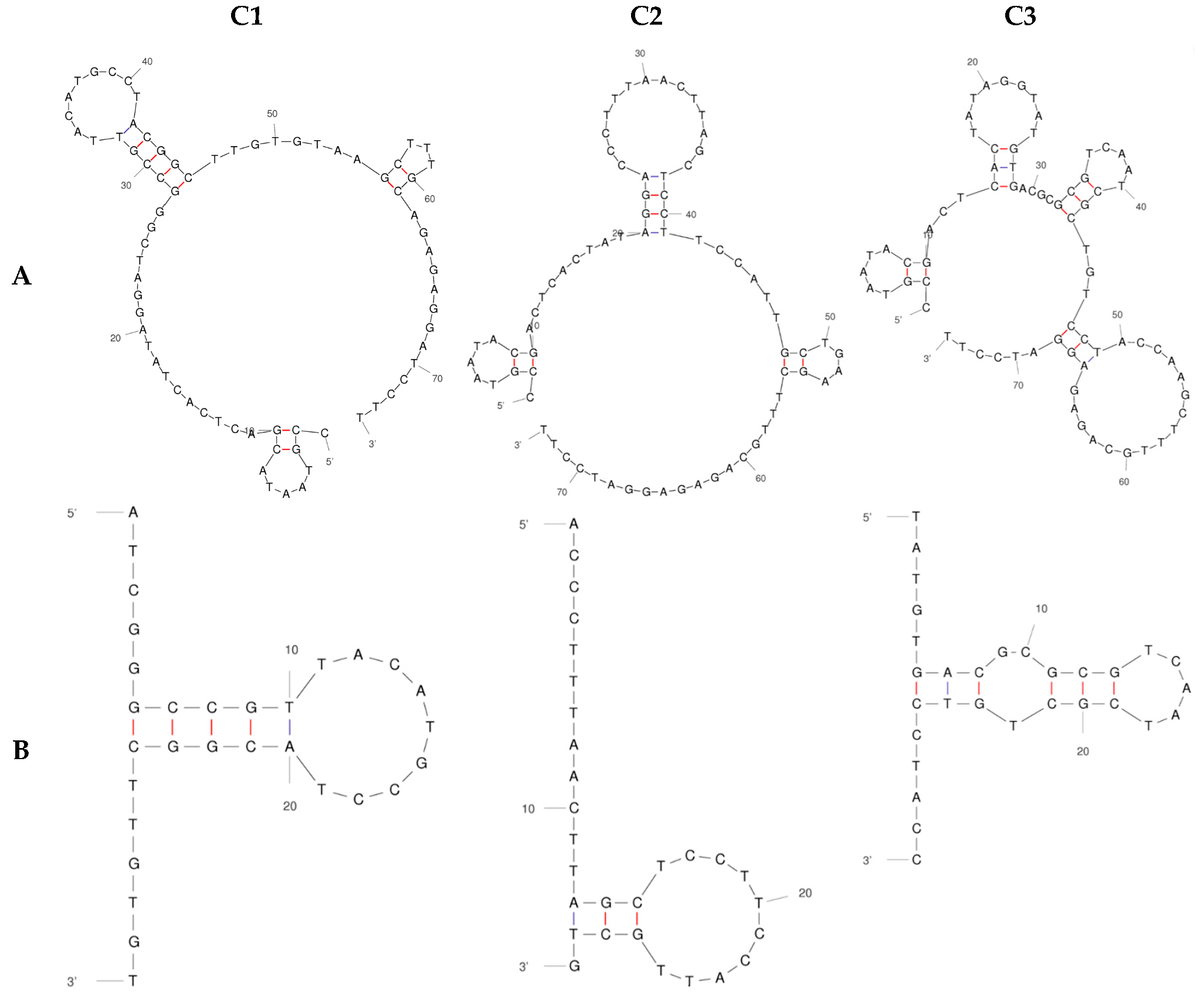
2.1. Strategy to Select DNA Aptamers against HECT Domain of WWP1
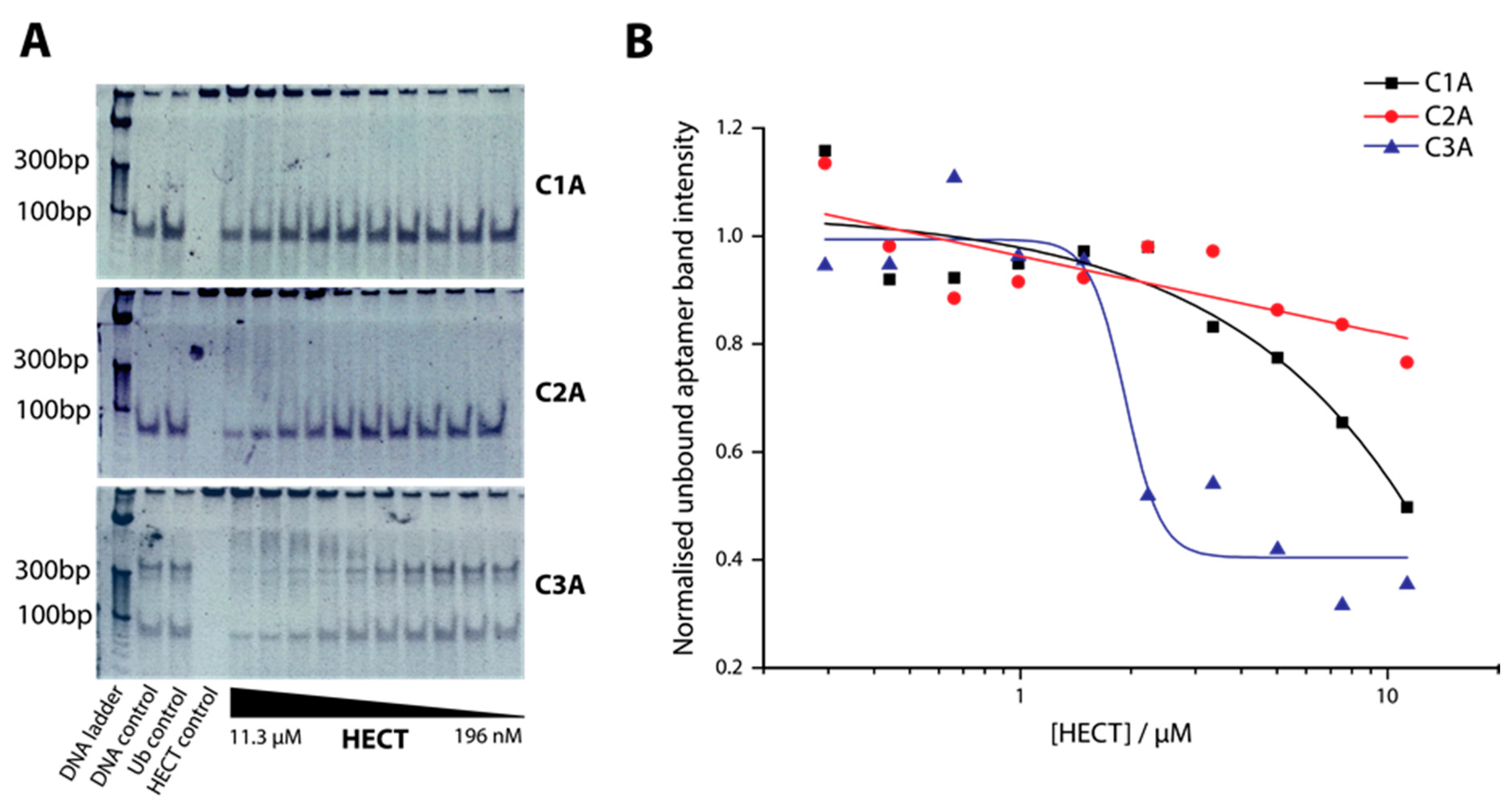
2.2. Determination of Binding Affinity of DNA Aptamers Binding to HECT Domain
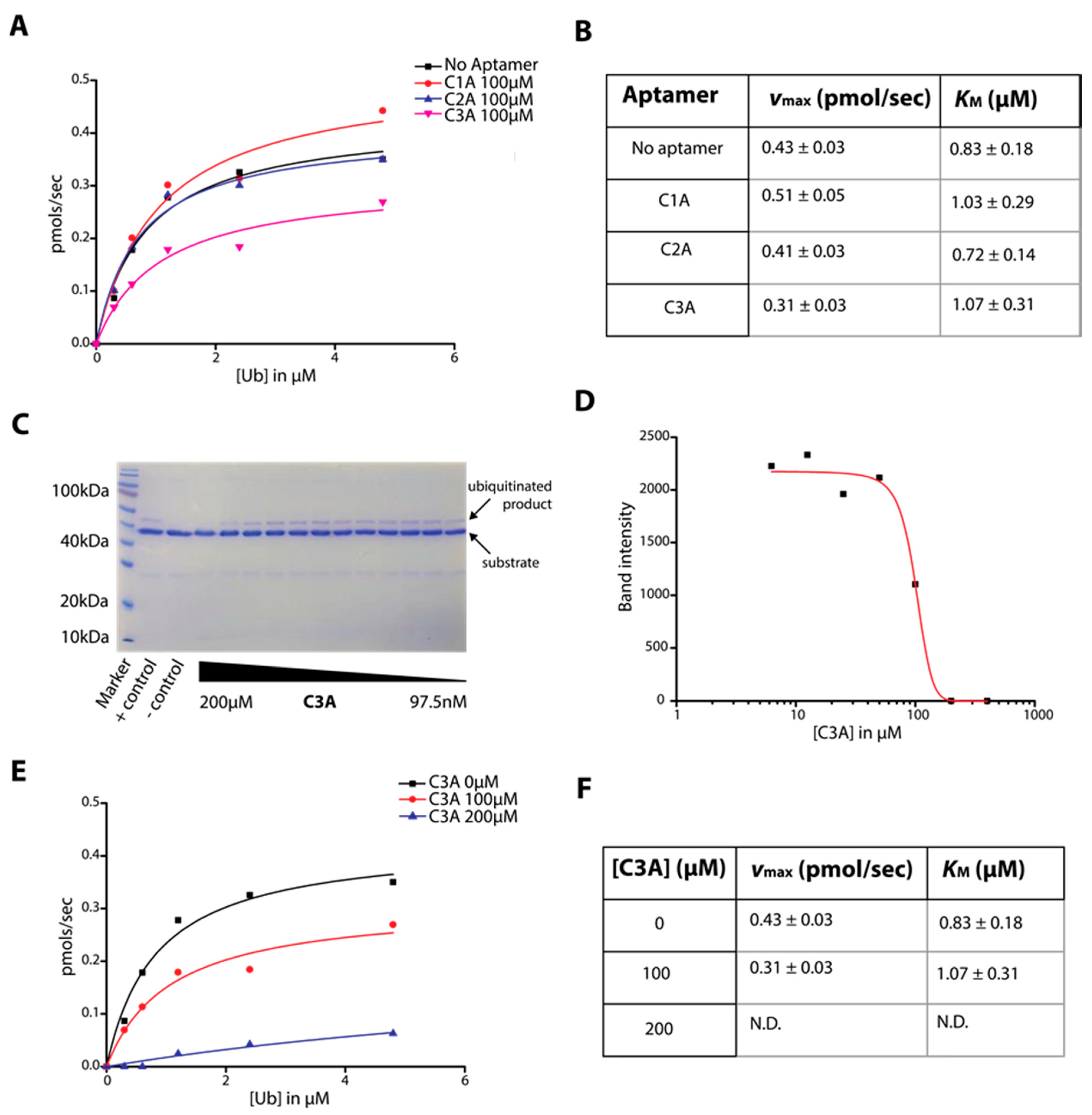
2.3. Determination of C-Lobe DNA Aptamer-Mediated Inhibition of Ubiquitination Activity of HECT Domain
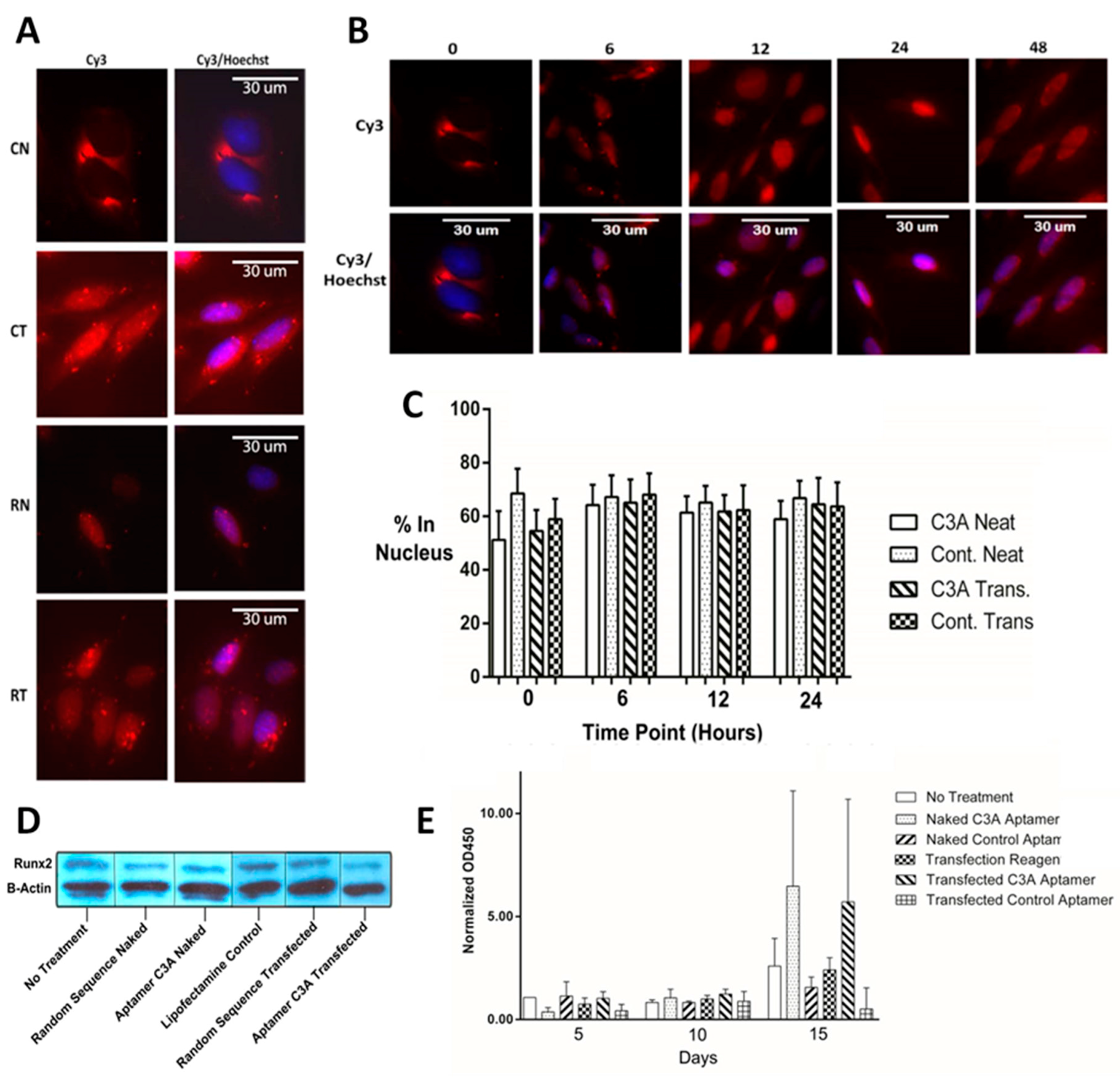
2.4. Cellular Localization, Runx2 Levels, and Bone Mineralization in Saos-2 Osteoblastic Cells
3. Discussion
4. Materials and Methods
4.1. HECT, C-Lobe, N-Lobe and E2 Cloning, Expression and Purification
4.2. SELEX Procedures
4.3. HECT Ubiquitin Ligase Activity Assay
4.4. Electrophoretic Mobility Shift Assay (EMSA)
4.5. Cell Culturing
4.6. Fixed Cell Imaging
4.7. Runx2 Western Blot
4.8. Apoptosis Assay
4.9. Bone Mineralization Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dallas, S.L.; Bonewald, L.F. Dynamics of the transition from osteoblast to osteocyte. Ann. N. Y. Acad. Sci. 2010, 1192, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Glimcher, L.H.; Jones, D.C.; Wein, M.N. Control of postnatal bone mass by the zinc finger adapter protein Schnurri-3. Ann. N. Y. Acad. Sci. 2007, 1116, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.H.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Wein, M.N.; Jones, D.C.; Shim, J.H.; Aliprantis, A.O.; Sulyanto, R.; Lazarevic, V.; Poliachik, S.L.; Gross, T.S.; Glimcher, L.H. Control of bone resorption in mice by Schnurri-3. Proc. Natl. Acad. Sci. USA 2012, 109, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.C.; Wein, M.N.; Oukka, M.; Hofstaetter, J.G.; Glimcher, M.J.; Glimcher, L.H. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science 2006, 312, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Johnson, K.D.; Petcherski, A.G.; Vandergon, T.; Mosser, E.A.; Copeland, N.G.; Jenkins, N.A.; Kimble, J.; Bresnick, E.H. A HECT domain ubiquitin ligase closely related to the mammalian protein WWP1 is essential for Caenorhabditis elegans embryogenesis. Gene 2000, 252, 137–145. [Google Scholar] [CrossRef]
- Pirozzi, G.; McConnell, S.J.; Uveges, A.J.; Carter, J.M.; Sparks, A.B.; Kay, B.K.; Fowlkes, D.M. Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem. 1997, 272, 14611–14616. [Google Scholar] [CrossRef] [PubMed]
- Verdecia, M.A.; Joazeiro, C.A.; Wells, N.J.; Ferrer, J.L.; Bowman, M.E.; Hunter, T.; Noel, J.P. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 2003, 11, 249–259. [Google Scholar] [CrossRef]
- Roth, A.; Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009, 78, 305–334. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.W.; Fraser, L.A.; Shiu, S.C.; Tang, M.S.; Tanner, J.A. Aptamer affinity maturation by resampling and microarray selection. Anal. Chem. 2016, 88, 6981–6985. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T. Aptamers and SELEX: The technology. World Pat. Inf. 2003, 25, 123–129. [Google Scholar] [CrossRef]
- Ulrich, H. DNA and RNA aptamers as modulators of protein function. Med. Chem. 2005, 1, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Cheung, Y.W.; Kwok, J.; Law, A.W.; Watt, R.M.; Kotaka, M.; Tanner, J.A. Structural basis for discriminatory recognition of plasmodium lactate dehydrogenase by a DNA aptamer. Proc. Natl. Acad. Sci. USA 2013, 110, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Huang, X.; Wu, L.; Chen, G.; Dong, J.; Cui, X.; Tang, Z. Selection of intracellularly functional RNA mimics of green fluorescent protein using fluorescence-activated cell sorting. J. Mol. Evol. 2015, 81, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Daniels, D.A.; Chen, H.; Hicke, B.J.; Swiderek, K.M.; Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci. USA 2003, 100, 15416–15421. [Google Scholar] [CrossRef] [PubMed]
- Stein, C.A.; Castanotto, D. FDA-approved oligonucleotide therapies in 2017. Mol. Ther. 2017, 25, 1069–1075. [Google Scholar] [CrossRef] [PubMed]
- Shum, K.T.; Chan, C.; Leung, C.M.; Tanner, J.A. Identification of a DNA aptamer that inhibits sclerostin’s antagonistic effect on Wnt signalling. Biochem. J. 2011, 434, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Dirkzwager, R.M.; Liang, S.; Tanner, J.A. Development of aptamer-based point-of-care diagnostic device for malaria using 3D printing rapid prototyping. ACS Sens. 2016, 1, 420–426. [Google Scholar] [CrossRef]
- Cheung, Y.W.; Dirkzwager, R.M.; Wong, W.C.; Cardoso, J.; Costa, J.D.; Tanner, J.A. Aptamer-mediated plasmodium-specific diagnosis of malaria. Biochimie 2018, 145, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Fraser, L.A.; Kinghorn, A.B.; Dirkzwager, R.M.; Liang, S.; Cheung, Y.W.; Lim, B.; Shiu, S.C.; Tang, M.S.L.; Andrew, D.; Manitta, J.; et al. A portable microfluidic Aptamer-Tethered Enzyme Capture (APTEC) biosensor for malaria diagnosis. Biosens. Bioelectron. 2018, 100, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Ouellet, J. RNA fluorescence with light-up aptamers. Front. Chem. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. Biotechnological mass production of DNA origami. Nature 2017, 552, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Maurisse, R.; De Semir, D.; Emamekhoo, H.; Bedayat, B.; Abdolmohammadi, A.; Parsi, H.; Gruenert, D.C. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 2010, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Zhao, X.; Lee, L.J.; Lee, R.J. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 2009, 11, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Zamecnik, P.C.; Stephenson, M.L. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.; Juliano, R.L. Cellular uptake and intracellular fate of antisense oligonucleotides. Trends Cell Biol. 1992, 2, 139–144. [Google Scholar] [CrossRef]
- Bennett, R.M. As nature intended? The uptake of DNA and oligonucleotides by eukaryotic cells. Antisense Res. Dev. 1993, 3, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Loke, S.L.; Stein, C.A.; Zhang, X.H.; Mori, K.; Nakanishi, M.; Subasinghe, C.; Cohen, J.S.; Neckers, L.M. Characterization of oligonucleotide transport into living cells. Proc. Natl. Acad. Sci. USA 1989, 86, 3474–3478. [Google Scholar] [CrossRef] [PubMed]
- Yakubov, L.A.; Deeva, E.A.; Zarytova, V.F.; Ivanova, E.M.; Ryte, A.S.; Yurchenko, L.V.; Vlassov, V.V. Mechanism of oligonucleotide uptake by cells: Involvement of specific receptors? Proc. Natl. Acad. Sci. USA 1989, 86, 6454–6458. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J.J. Therapeutic potential of aptamer-siRNA conjugates for treatment of HIV-1. BioDrugs 2012, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Legendre, J.Y.; Szoka, F.C., Jr. Delivery of plasmid DNA into mammalian cell lines using pH-sensitive liposomes: Comparison with cationic liposomes. Pharm. Res. 1992, 9, 1235–1242. [Google Scholar] [CrossRef] [PubMed]
- De Fougerolles, A.R. Delivery vehicles for small interfering RNA in vivo. Hum. Gene Ther. 2008, 19, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G. The chemical biology of aptamers. Angew. Chem. 2009, 48, 2672–2689. [Google Scholar] [CrossRef] [PubMed]
- Auslander, D.; Wieland, M.; Auslander, S.; Tigges, M.; Fussenegger, M. Rational design of a small molecule-responsive intramer controlling transgene expression in mammalian cells. Nucleic Acids Res. 2011, 39, e155. [Google Scholar] [CrossRef] [PubMed]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Matta-Camacho, E.; Kozlov, G.; Menade, M.; Gehring, K. Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol. Ther. 2003, 2, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Beaudenon, S.; Dastur, A.; Huibregtse, J.M. Expression and assay of HECT domain ligases. Methods Enzymol. 2005, 398, 112–125. [Google Scholar] [PubMed]
- Hu, J.; Wu, J.; Li, C.; Zhu, L.; Zhang, W.Y.; Kong, G.; Lu, Z.; Yang, C.J. A G-quadruplex aptamer inhibits the phosphatase activity of oncogenic protein Shp2 in vitro. Chembiochem 2011, 12, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ominsky, M.S.; Warmington, K.S.; Morony, S.; Gong, J.; Cao, J.; Gao, Y.; Shalhoub, V.; Tipton, B.; Haldankar, R.; et al. Sclerostin antibody treatment increases bone formation, bone mass, and bone strength in a rat model of postmenopausal osteoporosis. J. Bone Miner. Res. 2009, 24, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.D.; Rhodes, D.G.; Burgess, D.J. DNA-based therapeutics and DNA delivery systems: A comprehensive review. AAPS J. 2005, 7, E61–E77. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Reyes, E.; Šalipur, F.R.; Shams, M.; Forsthoefel, M.K.; Bates, P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015, 9, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Wente, S.R.; Rout, M.P. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2010, 2, a000562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wu, Z.; Ma, Z.; Liu, H.; Wu, Y.; Zhang, Q. WWP1 as a potential tumor oncogene regulates PTEN-Akt signaling pathway in human gastric carcinoma. Tumour. Biol. 2015, 36, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Kinghorn, A.B.; Fraser, L.A.; Lang, S.; Shiu, S.C.C.; Tanner, J.A. Aptamer bioinformatics. Int. J. Mol. Sci. 2017, 18, 2516. [Google Scholar] [CrossRef]
- Fraser, L.A.; Kinghorn, A.B.; Tang, M.S.; Cheung, Y.-W.; Lim, B.; Liang, S.; Dirkzwager, R.M.; Tanner, J.A. Oligonucleotide functionalised microbeads: Indispensable tools for high-throughput aptamer selection. Molecules 2015, 20, 21298–21312. [Google Scholar] [CrossRef] [PubMed] [Green Version]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tucker, W.O.; Kinghorn, A.B.; Fraser, L.A.; Cheung, Y.-W.; Tanner, J.A. Selection and Characterization of a DNA Aptamer Specifically Targeting Human HECT Ubiquitin Ligase WWP1. Int. J. Mol. Sci. 2018, 19, 763. https://doi.org/10.3390/ijms19030763
Tucker WO, Kinghorn AB, Fraser LA, Cheung Y-W, Tanner JA. Selection and Characterization of a DNA Aptamer Specifically Targeting Human HECT Ubiquitin Ligase WWP1. International Journal of Molecular Sciences. 2018; 19(3):763. https://doi.org/10.3390/ijms19030763
Chicago/Turabian StyleTucker, Wesley O., Andrew B. Kinghorn, Lewis A. Fraser, Yee-Wai Cheung, and Julian A. Tanner. 2018. "Selection and Characterization of a DNA Aptamer Specifically Targeting Human HECT Ubiquitin Ligase WWP1" International Journal of Molecular Sciences 19, no. 3: 763. https://doi.org/10.3390/ijms19030763