Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization
Abstract
:1. Introduction
2. Results
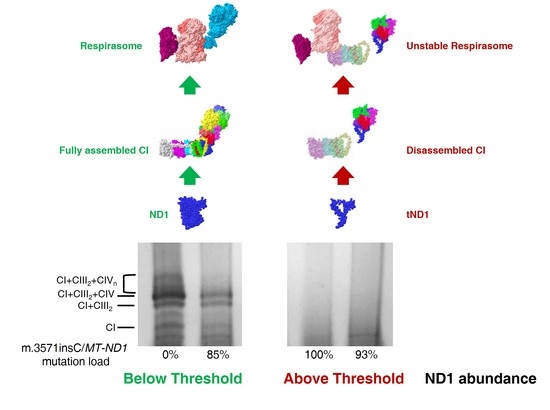
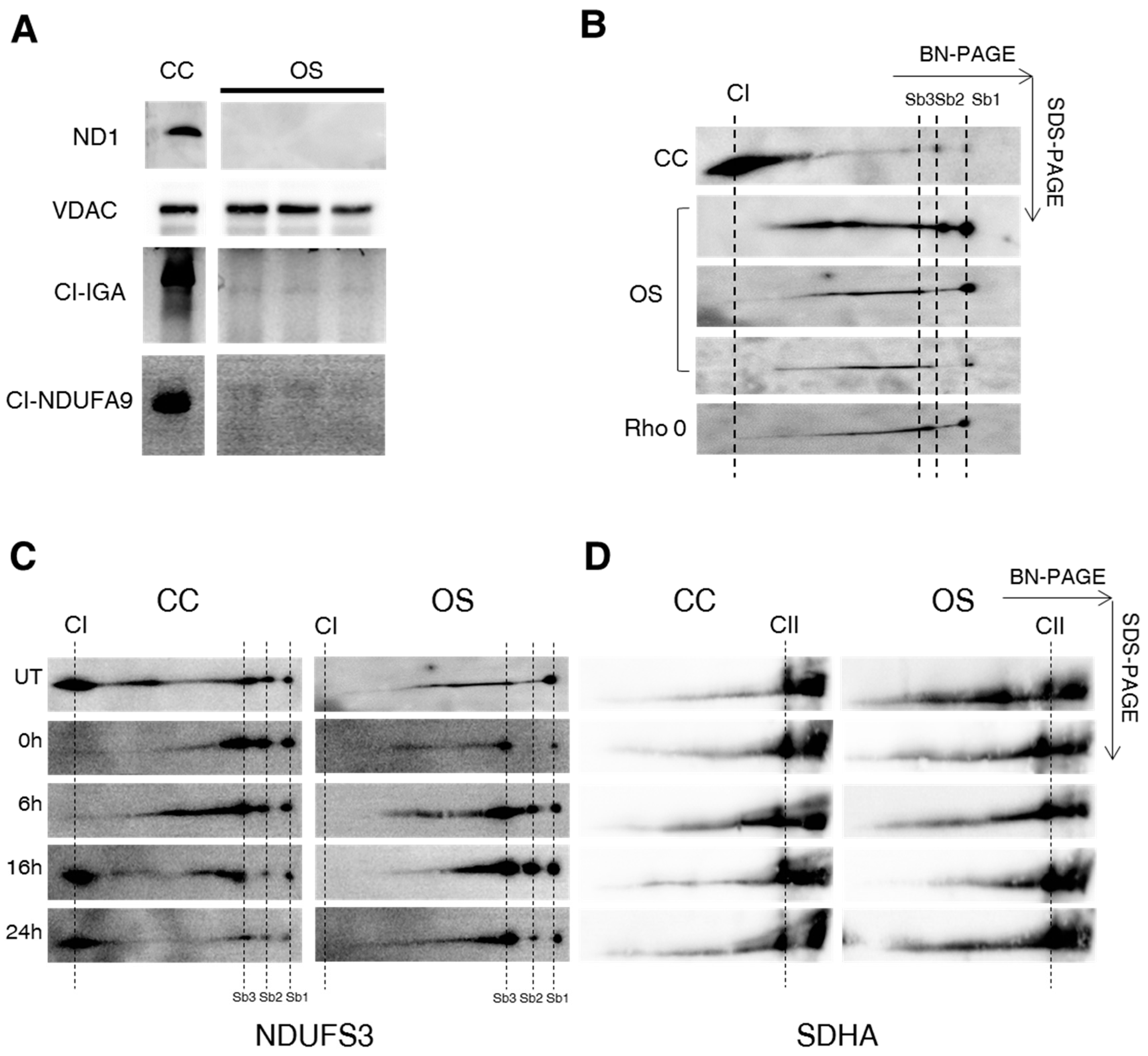
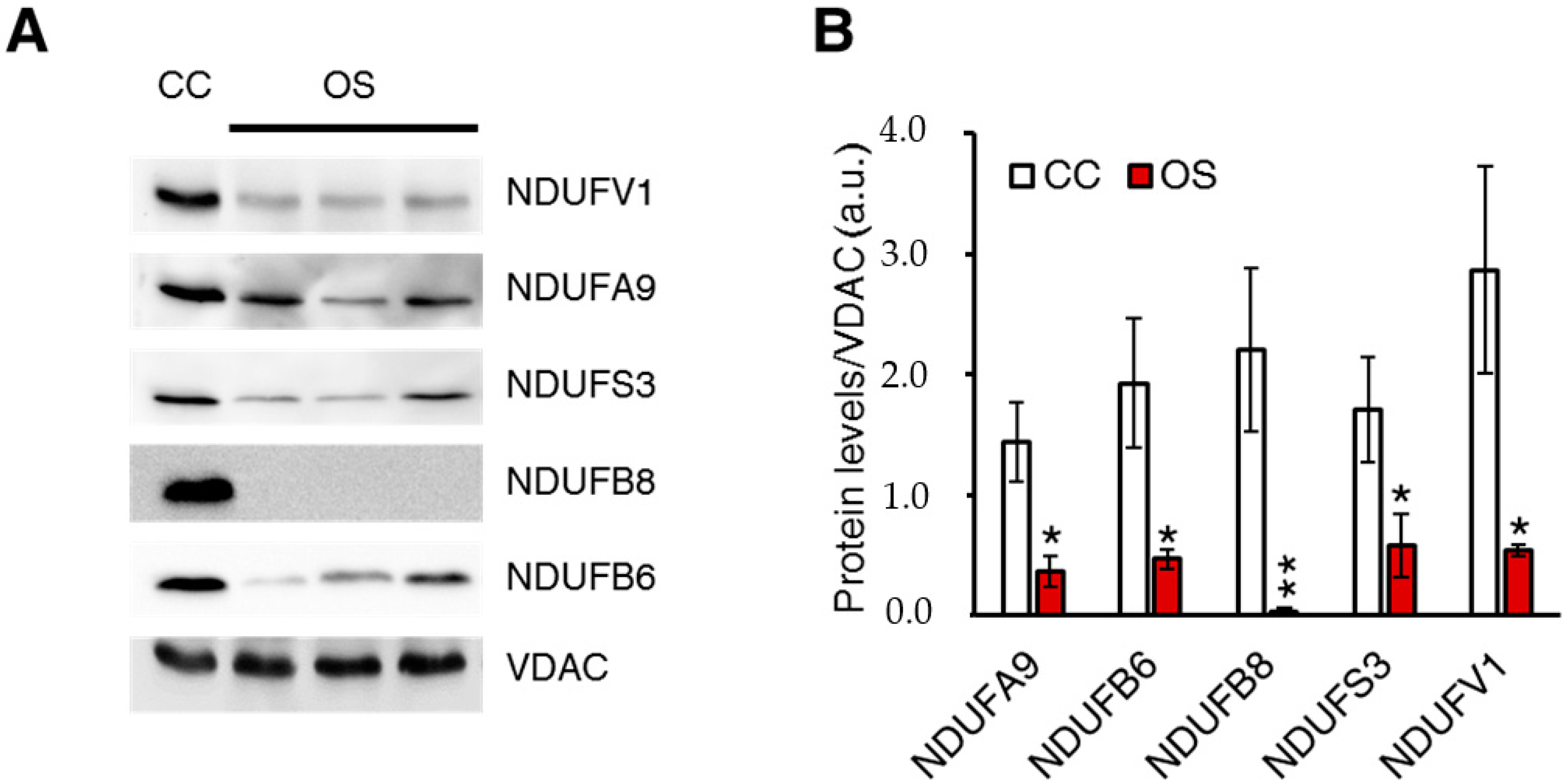
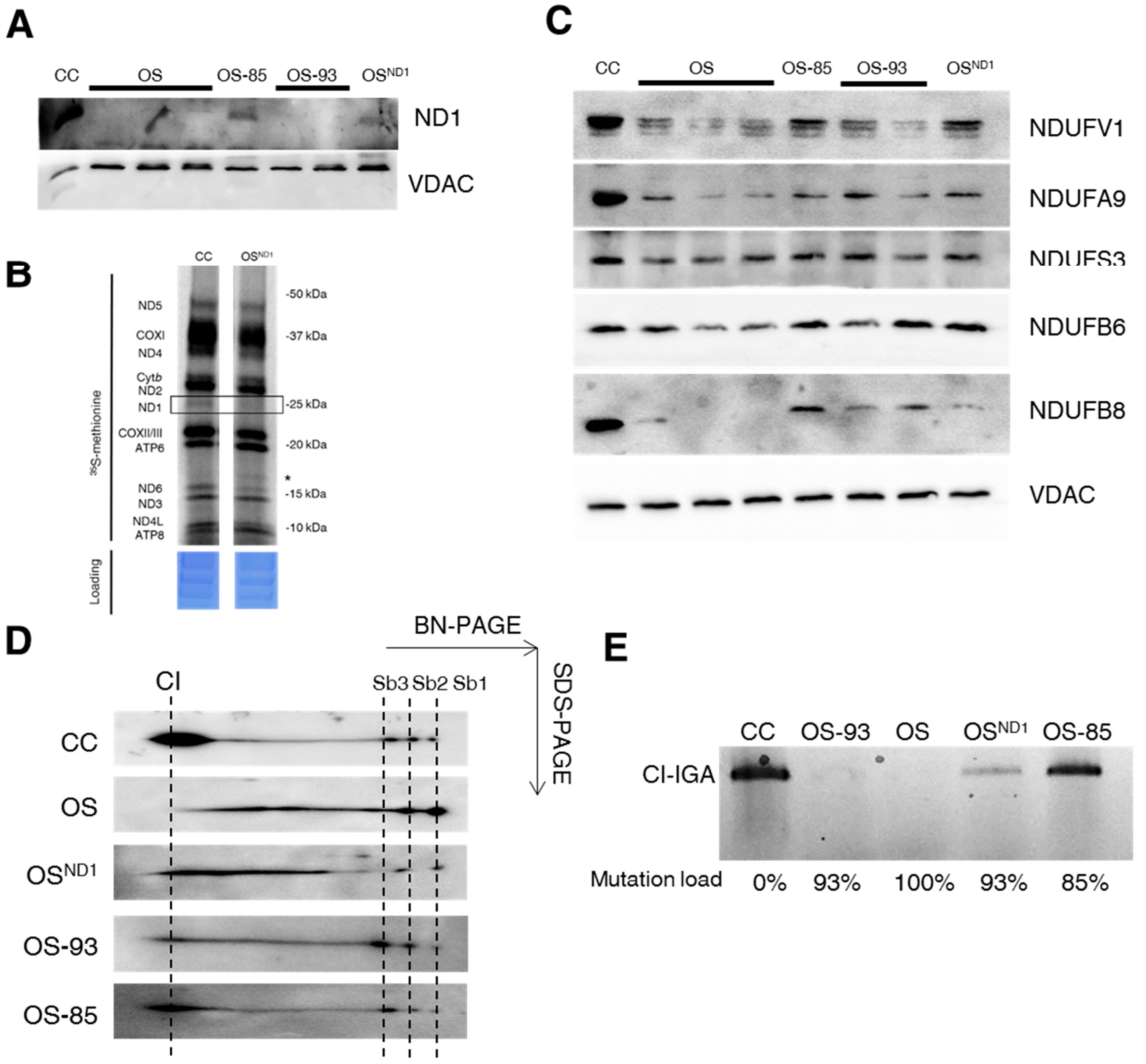
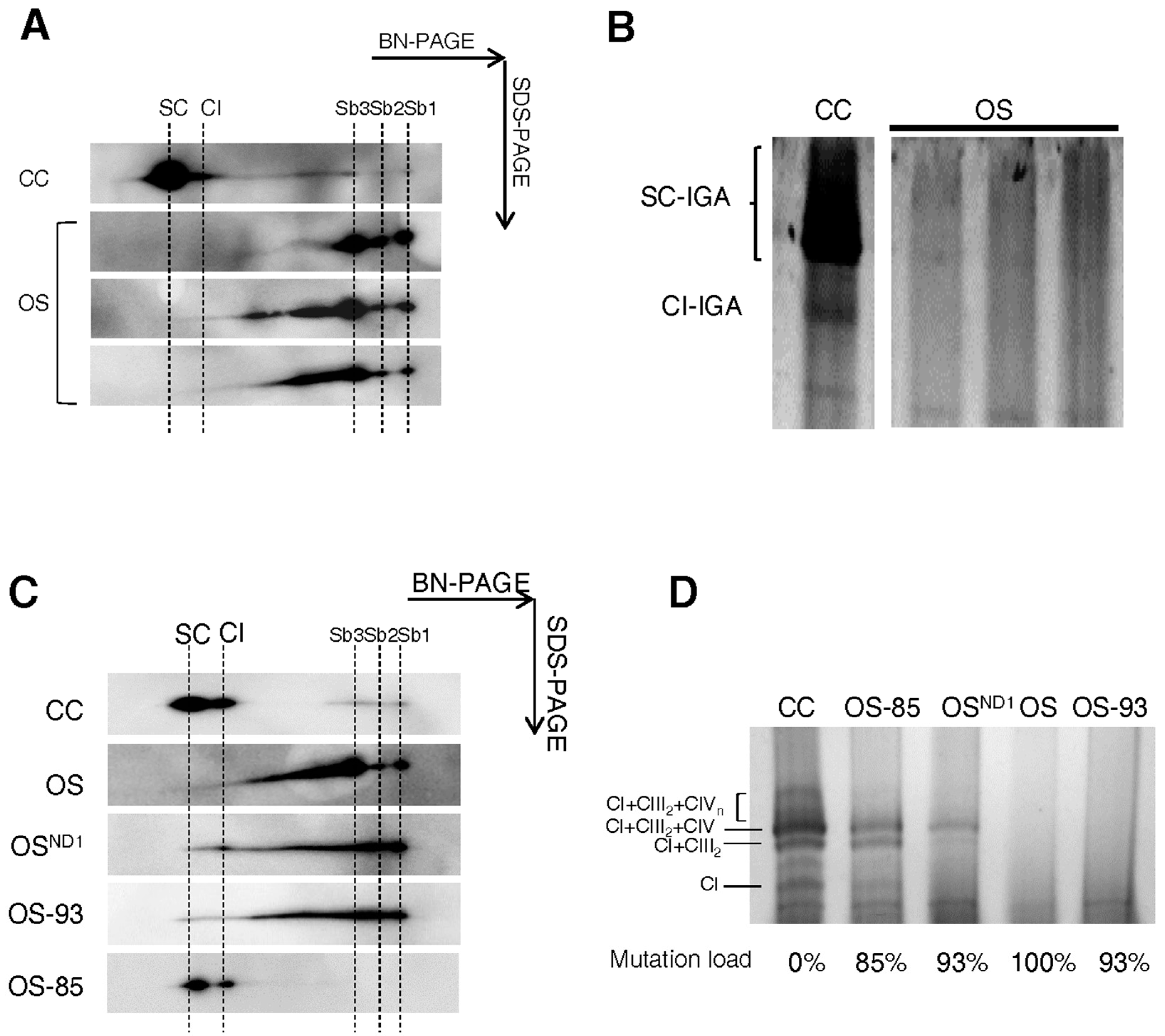
2.1. Ablation of ND1 Induced by Homoplasmic m.3571insC/MT-ND1 Mutation Determines a Stall in Complex I Assembly
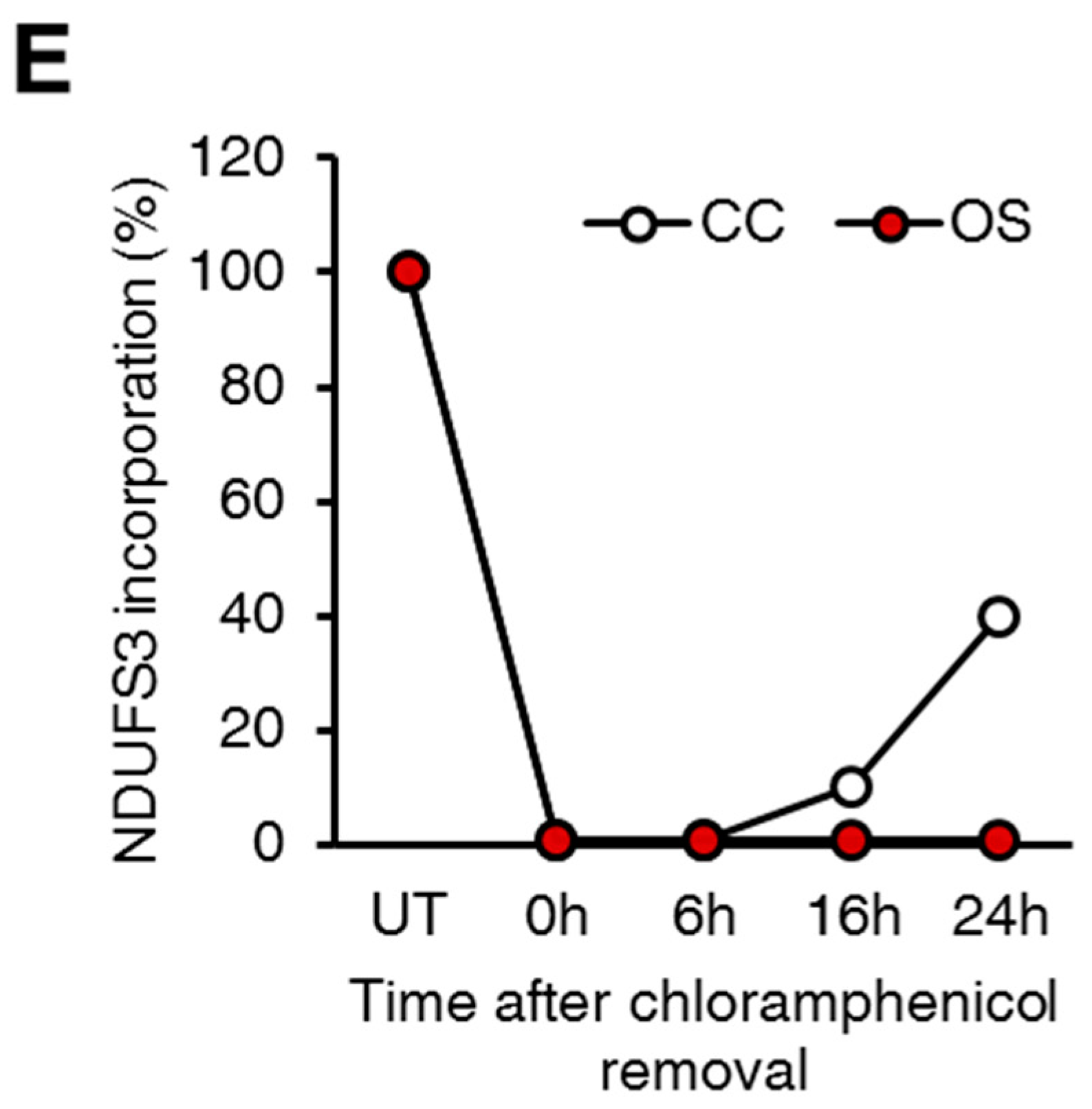
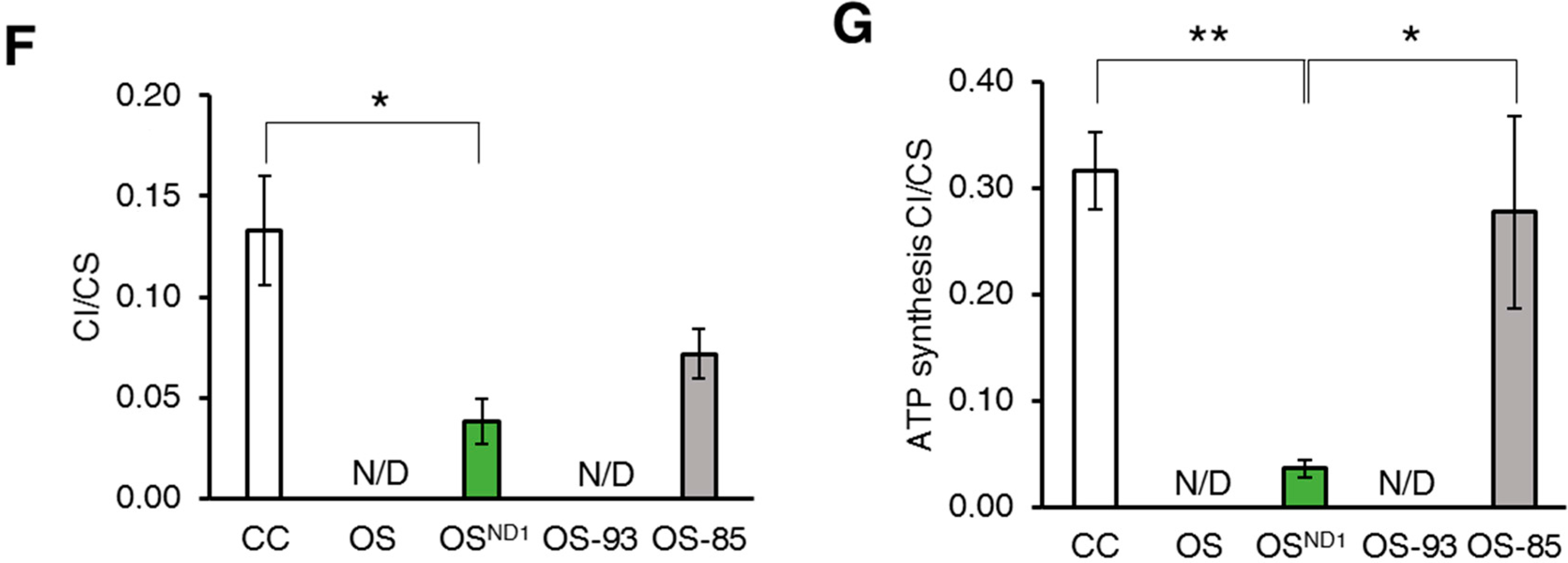
2.2. Small Amounts of Wild Type ND1 Subunit Are Sufficient to Partially Recover CI Disruption
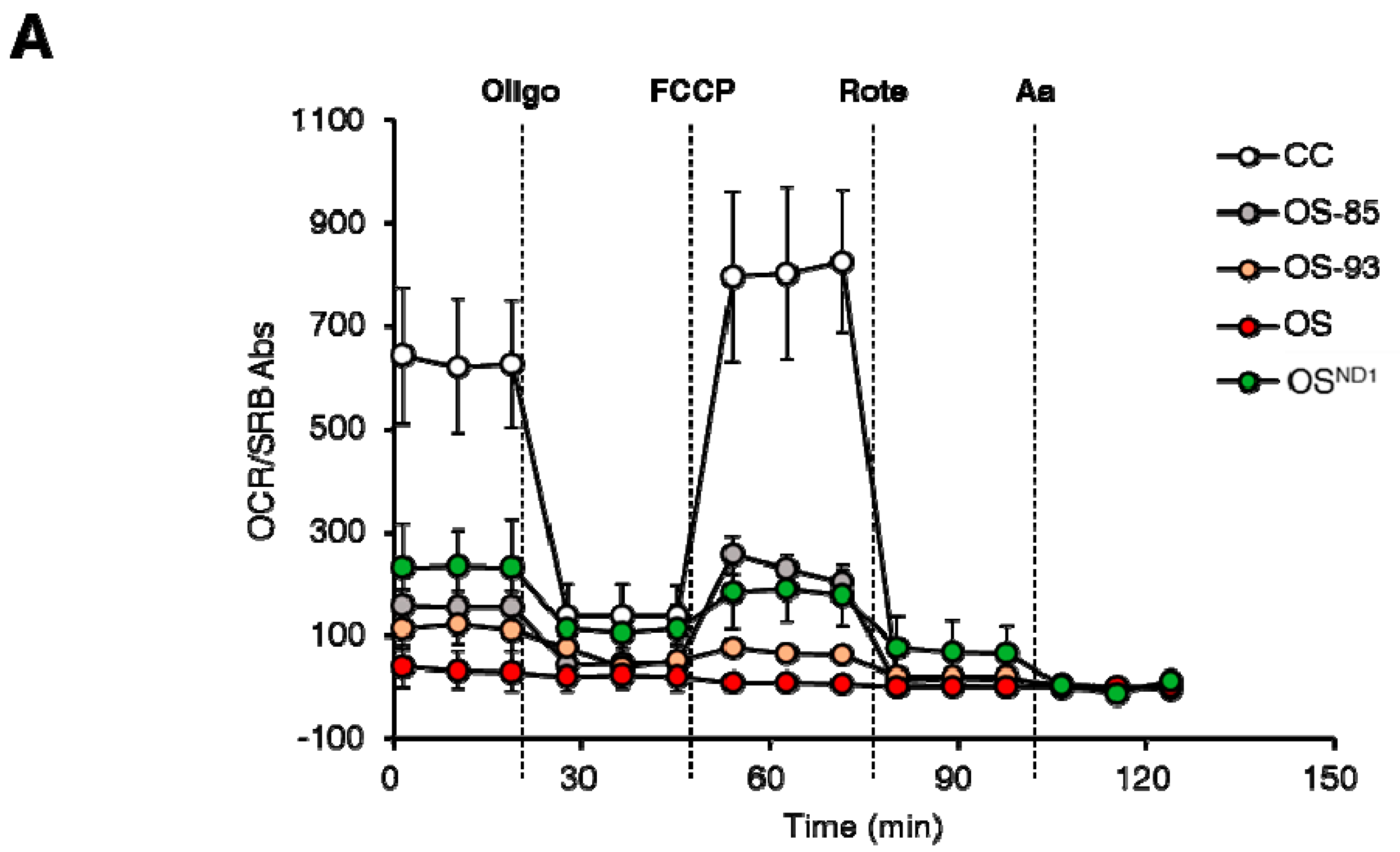
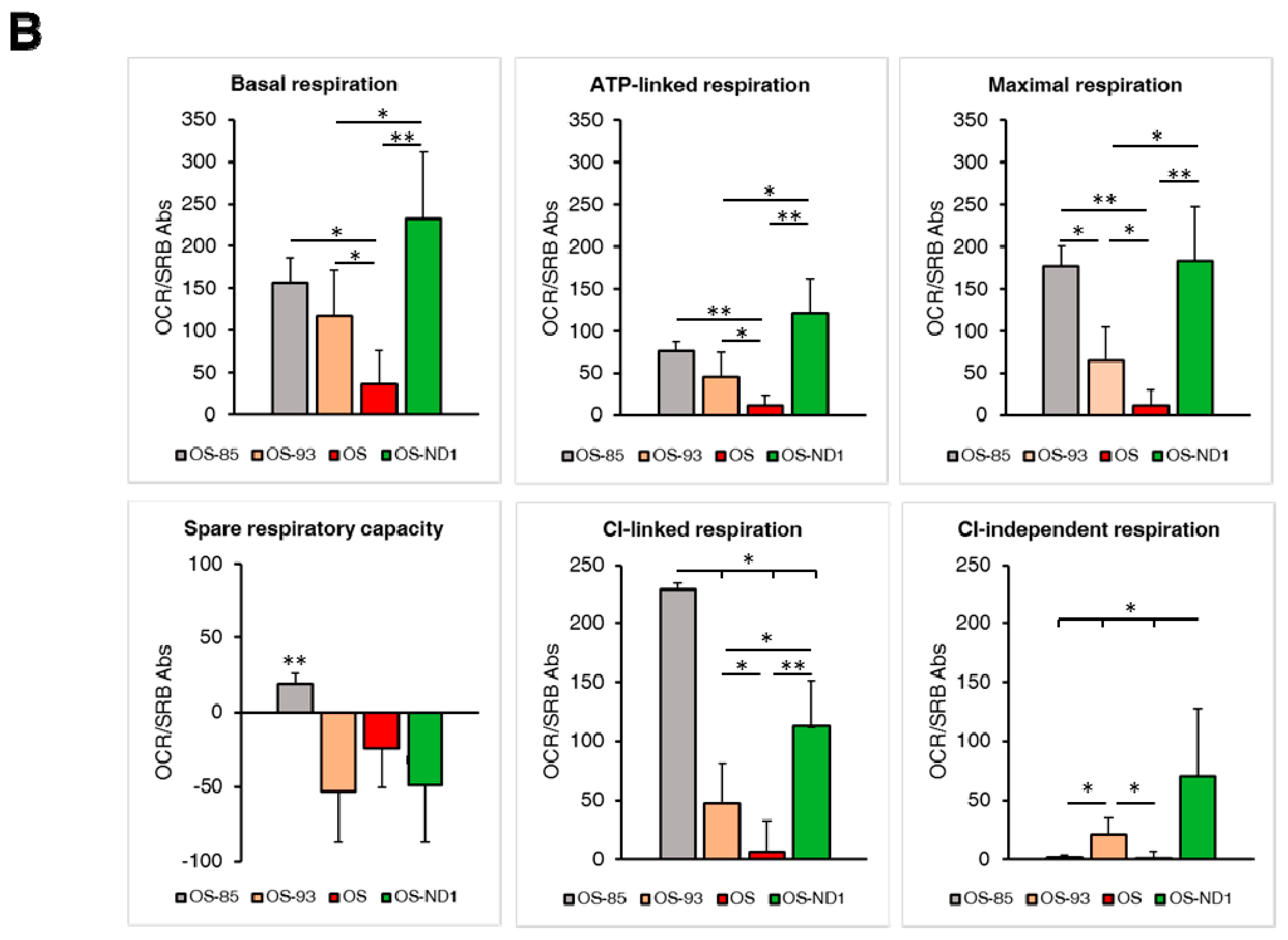
2.3. Lack of ND1 Hampers Supercomplexes Formation and Stability
3. Discussion
4. Methods
4.1. Cell Lines and Culture Conditions
4.2. Mitochondrial DNA Sequencing and Low Heteroplasmy Detection
4.3. Mitochondrial-Enriched Fraction and Crude Mitochondria Preparation
4.4. SDS-PAGE and Western Blot Analysis
4.5. High-Resolution Clear Native PAGE (hrCNE)
4.6. First Dimension Blue Native-PAGE (1D BN-PAGE)
4.7. 2D BN/SDS-PAGE
4.8. Complexes Re-Assembly Kinetics Assay
4.9. In Vitro Mitochondrial Translation Assay
4.10. Spectrophotometric Assays of Respiratory Complexes Activity
4.11. Mitochondrial ATP Synthesis
4.12. Microrespirometry
4.13. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 1D BN-PAGE | 1 dimension Blue Native PolyAcrylamide Gel Electrophoresis |
| 2D SDS/BN-PAGE | 2 dimension SDS/Blue Native PolyAcrylamide Gel Electrophoresis |
| ADP | Adenosine diphosphate |
| AMP | Adenosine monophosphate |
| AMPK | AMP-activated kinase |
| ATP | Adenosine triphosphate |
| BSA | Bovine Serum Albumine |
| CI | Complex I |
| CII | Complex II |
| CI-IGA | Complex I In-gel Activity |
| CIII | Complex III |
| CIV | Complex IV |
| CS | Citrate Synthase |
| CV | Complex V |
| DB | Decylubiquinone |
| DCIP | 2,6-dichlorophenolindophenol |
| DDM | n-dodecyl β-maltoside |
| DHPLC | Denaturing High Performance Liquid Chromatography |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EDTA | Ethylenediaminetetraacetic acid |
| EGTA | Ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid |
| F-PCR | Fluorescent PCR |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HIF-1α | Hypoxia Inducible factor 1α |
| hrCNE | High resolution Clear Native Electrophoresis |
| mtDNA | Mitochondrial DNA |
| NADH | Reduced nicotinamide adenine dinucleotide |
| nDNA | Nuclear DNA |
| OS | Osteosarcoma |
| OXPHOS | Oxidative phosphorylation |
| PBS | Phosphate buffered saline |
| Sb | Subcomplex |
| SDS-PAGE | Sodium dodecyl sulfate PolyAcrylamide Gel Electrophoresis |
References
- Hirst, J. Mitochondrial Complex I. Annu. Rev. Biochem. 2013, 82, 551–575. [Google Scholar] [CrossRef] [PubMed]
- Vinothkumar, K.R.; Zhu, J.; Hirst, J. Architecture of mammalian respiratory complex I. Nature 2014, 515, 80–84. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Castillo, S.; Baertling, F.; Kownatzki, D.; Wessels, H.J.; Arnold, S.; Brandt, U.; Nijtmans, L. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell Metab. 2017, 25, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Caballero, L.; Guerrero-Castillo, S.; Nijtmans, L. Unraveling the complexity of mitochondrial complex I assembly: A dynamic process. Biochim. Biophys. Acta 2016. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, R.; Berrisford, J.M.; Minhas, G.S.; Sazanov, L.A. Crystal structure of the entire respiratory complex I. Nature 2013, 494, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Fiedorczuk, K.; Letts, J.A.; Degliesposti, G.; Kaszuba, K.; Skehel, M.; Sazanov, L.A. Atomic structure of the entire mammalian mitochondrial complex I. Nature 2016, 538, 406–410. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Vinothkumar, K.R.; Hirst, J. Structure of mammalian respiratory complex I. Nature 2016, 536, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Zickermann, V.; Wirth, C.; Nasiri, H.; Siegmund, K.; Schwalbe, H.; Hunte, C.; Brandt, U. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science 2015, 347, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Letts, J.A.; Fiedorczuk, K.; Sazanov, L.A. The architecture of respiratory supercomplexes. Nature 2016, 537, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Lastres, D.; Fontanesi, F.; García-Consuegra, I.; Martín, M.A.; Arenas, J.; Barrientos, A.; Ugalde, C. Mitochondrial Complex I Plays an Essential Role in Human Respirasome Assembly. Cell Metab. 2012, 15, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; Pfeiffer, K. The ratio of oxidative phosphorylation complexes I, II, III, IV, and V in bovine heart mitochondria, and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001. [Google Scholar] [CrossRef]
- Wu, M.; Gu, J.; Guo, R.; Huang, Y.; Yang, M. Structure of Mammalian Respiratory Supercomplex I1III2IV1. Cell 2016, 167, 1598–1609.e10. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Zong, S.; Wu, M.; Gu, J.; Yang, M. Architecture of Human Mitochondrial Respiratory Megacomplex I2III2IV2. Cell 2017, 170, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Blaza, J.N.; Larsson, N.-G.; Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metab. 2017, 25, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Iommarini, L.; Calvaruso, M.A.; Kurelac, I.; Gasparre, G.; Porcelli, A.M. Complex I impairment in mitochondrial diseases and cancer: Parallel roads leading to different outcomes. Int. J. Biochem. Cell Biol. 2013, 45, 47–63. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Chinnery, P.F. The dynamics of mitochondrial DNA heteroplasmy: Implications for human health and disease. Nat. Rev. Genet. 2015, 16, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, R.; Faustin, B.; Rocher, C.; Malgat, M.; Mazat, J.-P.; Letellier, T. Mitochondrial threshold effects. Biochem. J. 2003, 370, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Bonora, E.; Porcelli, A.M.; Gasparre, G.; Biondi, A.; Ghelli, A.; Carelli, V.; Baracca, A.; Tallini, G.; Martinuzzi, A.; Lenaz, G.; et al. Defective oxidative phosphorylation in thyroid oncocytic carcinoma is associated with pathogenic mitochondrial DNA mutations affecting complexes I and III. Cancer Res. 2006, 66, 6087–6096. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.O.; Dieteren, C.E.J.; van den Heuvel, L.P.W.J.; Willems, P.H.G.M.; Smeitink, J.A.M.; Koopman, W.J.H.; Nijtmans, L.G.J. Identification of mitochondrial complex I assembly intermediates by tracing tagged NDUFS3 demonstrates the entry point of mitochondrial subunits. J. Biol. Chem. 2007, 282, 7582–7590. [Google Scholar] [CrossRef] [PubMed]
- Kurelac, I.; Lang, M.; Zuntini, R.; Calabrese, C.; Simone, D.; Vicario, S.; Santamaria, M.; Attimonelli, M.; Romeo, G.; Gasparre, G. Searching for a needle in the haystack: Comparing six methods to evaluate heteroplasmy in difficult sequence context. Syst. Biol. Biomed. Innov. 2012, 30, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Iommarini, L.; Kurelac, I.; Calvaruso, M.A.; Capristo, M.; Lollini, P.-L.; Nanni, P.; Bergamini, C.; Nicoletti, G.; Giovanni, C.D.; et al. Respiratory complex I is essential to induce a Warburg profile in mitochondria-defective tumor cells. Cancer Metab. 2013, 1, 11. [Google Scholar] [CrossRef] [PubMed]
- Lapuente-Brun, E.; Moreno-Loshuertos, R.; Acín-Pérez, R.; Latorre-Pellicer, A.; Colás, C.; Balsa, E.; Perales-Clemente, E.; Quirós, P.M.; Calvo, E.; Rodríguez-Hernández, M.A.; et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science 2013, 340, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Porcelli, A.M.; Ghelli, A.; Ceccarelli, C.; Lang, M.; Cenacchi, G.; Capristo, M.; Pennisi, L.F.; Morra, I.; Ciccarelli, E.; Melcarne, A.; et al. The genetic and metabolic signature of oncocytic transformation implicates HIF1alpha destabilization. Hum. Mol. Genet. 2010, 19, 1019–1032. [Google Scholar] [CrossRef] [PubMed]
- Gasparre, G.; Kurelac, I.; Capristo, M.; Iommarini, L.; Ghelli, A.; Ceccarelli, C.; Nicoletti, G.; Nanni, P.; de Giovanni, C.; Scotlandi, K.; et al. A mutation threshold distinguishes the antitumorigenic effects of the mitochondrial gene MTND1, an oncojanus function. Cancer Res. 2011, 71, 6220–6229. [Google Scholar] [CrossRef] [PubMed]
- Mayr, J.A.; Meierhofer, D.; Zimmermann, F.; Feichtinger, R.; Kögler, C.; Ratschek, M.; Schmeller, N.; Sperl, W.; Kofler, B. Loss of complex I due to mitochondrial DNA mutations in renal oncocytoma. Clin. Cancer Res. 2008, 14, 2270–2275. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, M.; Chiotis, M.; Hroudová, J.; Lopez Sanchez, M.I.G.; Lim, S.C.; Cook, M.J.; McKelvie, P.; Cotton, R.G.H.; Murphy, M.; St John, J.C.; et al. Capture of somatic mtDNA point mutations with severe effects on oxidative phosphorylation in synaptosome cybrid clones from human brain. Hum. Mutat. 2014, 35, 1476–1484. [Google Scholar] [CrossRef] [PubMed]
- Cardol, P.; Matagne, R.F.; Remacle, C. Impact of mutations affecting ND mitochondria-encoded subunits on the activity and assembly of complex I in chlamydomonas. implication for the structural organization of the enzyme. J. Mol. Biol. 2002, 319, 1211–1221. [Google Scholar] [CrossRef]
- Sinha, P.K.; Torres-Bacete, J.; Nakamaru-Ogiso, E.; Castro-Guerrero, N.; Matsuno-Yagi, A.; Yagi, T. Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J. Biol. Chem. 2009, 284, 9814–9823. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.C.; Hroudová, J.; Van Bergen, N.J.; Lopez Sanchez, M.I.G.; Trounce, I.A.; McKenzie, M. Loss of mitochondrial DNA-encoded protein ND1 results in disruption of complex I biogenesis during early stages of assembly. FASEB J. 2016. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, C.; Vogel, R.; Huijbens, R.; van den Heuvel, B.; Smeitink, J.; Nijtmans, L. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: A framework to interpret complex I deficiencies. Hum. Mol. Genet. 2004, 13, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Vartak, R.; Deng, J.; Fang, H.; Bai, Y. Redefining the roles of mitochondrial DNA-encoded subunits in respiratory complex I assembly. Biochim. Biophys. Acta 2015, 1852, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Kurelac, I.; MacKay, A.; Lambros, M.B.K.; di Cesare, E.; Cenacchi, G.; Ceccarelli, C.; Morra, I.; Melcarne, A.; Morandi, L.; Calabrese, F.M.; et al. Somatic complex I disruptive mitochondrial DNA mutations are modifiers of tumorigenesis that correlate with low genomic instability in pituitary adenomas. Hum. Mol. Genet. 2013, 22, 226–238. [Google Scholar] [CrossRef] [PubMed]
- Boulet, L.; Karpati, G.; Shoubridge, E.A. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 1992, 51, 1187–1200. [Google Scholar] [PubMed]
- Dunbar, D.R.; Moonie, P.A.; Zeviani, M.; Holt, I.J. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 1996, 5, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Miyabayashi, S.; Hanamizu, H.; Nakamura, R.; Endo, H.; Tada, K. Defects of mitochondrial respiratory enzymes in cloned cells from MELAS fibroblasts. J. Inherit. Metab. Dis. 1992, 15, 797–802. [Google Scholar] [CrossRef] [PubMed]
- Moslemi, A.R.; Tulinius, M.; Holme, E.; Oldfors, A. Threshold expression of the tRNA(Lys) A8344G mutation in single muscle fibres. Neuromuscul. Disord. 1998, 8, 345–349. [Google Scholar] [CrossRef]
- Fraidakis, M.J.; Jardel, C.; Allouche, S.; Nelson, I.; Auré, K.; Slama, A.; Lemière, I.; Thenint, J.P.; Hamon, J.B.; Zagnoli, F.; et al. Phenotypic diversity associated with the MT-TV gene m.1644G>A mutation, a matter of quantity. Mitochondrion 2014, 15, 34–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iommarini, L.; Kurelac, I.; Capristo, M.; Calvaruso, M.A.; Giorgio, V.; Bergamini, C.; Ghelli, A.; Nanni, P.; de Giovanni, C.; Carelli, V.; et al. Different mtDNA mutations modify tumor progression in dependence of the degree of respiratory complex I impairment. Hum. Mol. Genet. 2014, 23, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Acín-Pérez, R.; Fernández-Silva, P.; Peleato, M.L.; Pérez-Martos, A.; Enriquez, J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell 2008, 32, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Ghelli, A.; Tropeano, C.V.; Calvaruso, M.A.; Marchesini, A.; Iommarini, L.; Porcelli, A.M.; Zanna, C.; De Nardo, V.; Martinuzzi, A.; Wibrand, F.; et al. The cytochrome b p.278Y>C mutation causative of a multisystem disorder enhances superoxide production and alters supramolecular interactions of respiratory chain complexes. Hum. Mol. Genet. 2013, 22, 2141–2151. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lipton, J.H.; McMurray, W.C. Mitochondrial biogenesis in cultured animal cells I. Effect of chloramphenicol on morphology and mitochondrial respiratory enzymes. Biochim. Biophys. Acta 1977, 477, 264–272. [Google Scholar] [CrossRef]
| Cell Line | Mutation Load |
|---|---|
| CC | 0% |
| OS | 100% |
| OS-93 | 92.8 ± 0.3% |
| OS-85 | 85.3 ± 0.4% |
| OSND1 | 93.4 ± 1.3% |
| Rho 0 | no mtDNA |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iommarini, L.; Ghelli, A.; Tropeano, C.V.; Kurelac, I.; Leone, G.; Vidoni, S.; Lombes, A.; Zeviani, M.; Gasparre, G.; Porcelli, A.M. Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization. Int. J. Mol. Sci. 2018, 19, 764. https://doi.org/10.3390/ijms19030764
Iommarini L, Ghelli A, Tropeano CV, Kurelac I, Leone G, Vidoni S, Lombes A, Zeviani M, Gasparre G, Porcelli AM. Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization. International Journal of Molecular Sciences. 2018; 19(3):764. https://doi.org/10.3390/ijms19030764
Chicago/Turabian StyleIommarini, Luisa, Anna Ghelli, Concetta Valentina Tropeano, Ivana Kurelac, Giulia Leone, Sara Vidoni, Anne Lombes, Massimo Zeviani, Giuseppe Gasparre, and Anna Maria Porcelli. 2018. "Unravelling the Effects of the Mutation m.3571insC/MT-ND1 on Respiratory Complexes Structural Organization" International Journal of Molecular Sciences 19, no. 3: 764. https://doi.org/10.3390/ijms19030764