1. Introduction
Head and neck cancer is ranked as the seventh most frequent cause of cancer death in the world and squamous cell carcinoma comprises the most common subgroup [
1]. Despite ongoing advances in surgery and in radio- and chemotherapy, five-year survival rates for head and neck squamous cell carcinoma (HNSCC) remain still in the order of 50% to 60% [
1,
2]. Besides Human Papilloma Virus (HPV) status, prediction of clinical outcome and therapy are still based on histopathological and clinical parameters [
3,
4]. Novel therapeutic targets and markers to stratify patients are therefore urgently needed.
The NOTCH signaling pathway is becoming increasingly relevant in diverse tumor entities including HNSCC [
5,
6]. NOTCH signaling plays an integral part in cell fate and development by controlling proliferation, differentiation, angiogenesis, and apoptosis [
7]. Initiation of signaling is mediated through binding of the ligands Jagged or Delta-like resulting in the cleavage and release of NOTCH intracellular fragments (NOTCH-IC) [
5,
7,
8,
9]. Subsequently, NOTCH-IC are translocated to the nucleus and interact with RBPJ, a DNA-binding protein [
5,
7]. This leads to transcription of targets such as the MYC transcription factor and HES and HEY family proteins [
5,
7].
Alterations in NOTCH signaling have been described in several cancers and result in tumor promotion or suppression depending on the cancer entity and context [
5]. The predominant function of NOTCH signaling remains controversial—it may act as an oncogene, tumor suppressor, or even have a bimodal role [
10]. Frequent mutations of the NOTCH receptor family were detected in HNSCC and most likely result in loss of function of the receptors [
10,
11,
12]. A high incidence of nonsynonymous mutations was identified in 43% of Chinese patients with oral squamous cell carcinoma (OSCC) and the occurrence of mutations was associated with poor overall survival [
13,
14]. The expression of NOTCH1 pathway genes, however, has only been studied in small patient cohorts or in subgroups of HNSCC with contradictory results.
Currently, the clinical relevance of transcriptional alterations in the NOTCH signaling pathway in HNSCC is not well understood. The aim of this study was therefore to evaluate the expression of key components of the NOTCH pathway with quantitative real-time PCR in a larger HNSCC collective and in normal tissue. Secondly, the association of the expression with clinical and pathological parameters was investigated. Moreover, NOTCH1 expression was also analyzed in HPV-positive and -negative HNSCC cell lines.
4. Materials and Methods
4.1. Patient Tissue Samples
We obtained tissue samples from 195 HNSCC Patients (163 males, 32 females; median age 59 years, range 35 to 89 years) diagnosed between January 2002 and December 2005. As a control group we used mucosa obtained through panendoscopy or tonsillectomy (
n = 30, 18 males, 12 females; median age 51 years, range 25 to 87 years) from oropharynx (
n = 7), hypopharynx (
n = 17), and larynx (
n = 6). All patients were treated in the Department of Otorhinolaryngology at Klinikum rechts der Isar, Technical University of Munich. Sixty-five patients of the cohort have been used for a previous study [
9]. All tissue samples were formalin fixed and paraffin embedded (FFPE). Only one patient received neoadjuvant therapy; all remaining specimens were retrieved before adjuvant or primary radiochemotherapy. The independent ethics committee of the Technical University of Munich approved the study, under project number 1420/05 (13 June 2014) and 107/15 (12 March 2015).
4.2.Clinical Data
Clinical data provided by the Munich Cancer Registry were verified with data gathered from filed and electronical medical records. Thirty-nine patients (20.0%) were treated solely with surgery; 95 patients (48.7%) underwent surgical resection combined with radio(-chemo)therapy. Primary radiation or radio-chemotherapy was used in 48 patients (24.6%); 5 patients (2.6%) received palliative radio- and/or chemotherapy. In 8 cases (4.1%), no treatment was applied or treatment could not be reproduced. The median survival calculated by Kaplan-Meier analysis was 4.24 years (min–max follow up period 0.09–14.14 years); the overall five-year survival rate was 46.6%.
4.3. RNA Isolation and cDNA Synthesis
FFPE tissue samples were deparaffinized and digested using 40 µL Proteinase K (Roche Diagnostics GmbH, Unterhaching, Germany) in 100 µL PK buffer (50 mM Tris, 1 mM EDTA, and 25% Tween 20 diluted in water) added to 16 µL 10% SDS (10 g Sodiumdodecylsulfat diluted in 100 mL water) at 55 °C. After 24 h, 10 µL Proteinase K was added again and samples were incubated for another day. Further processing was performed using an InviTrap® RNA Mini Kit (Stratec, Birkenfeld, Germany) according to the manufacturer’s protocol. RNA from HNSCC cells was isolated with the RNeasy-Mini-Kit (Qiagen, Hilden, Germany). After isolation the RNA concentration was quantified using the NanoDrop 1000 system (PEQLAB, Erlangen, Germany). We used only probes with a minimal RNA concentration of 10 ng/µL. Afterwards, probes were diluted to a concentration of 10 or 25 ng/µL depending on the initial concentration of RNA and stored at −20 °C. cDNA synthesis was performed using Maxima® reverse transcriptase (Fermentas, Waltham, MA, USA) according to the manufacturer’s protocol.
4.4. Quantitative Real-Time PCR
Quantitative real-time PCR (qPCR) was performed to quantify mRNA expression of NOTCH1, NOTCH3, HES1, HEY1, JAG1, HPV16 E6, and HPV16 E7. For normalization of expression levels, GAPDH was used per sample. For qPCR mix, 50 ng cDNA template was added to 12.5 µL KAPA-SYBR Fast Universal (PeqLab, Erlangen, Germany) and 0.5 µL of 20 pmol of each primer. Water was added to a final volume of 25 µL. Primer sequences and specific annealing temperatures are depicted in
Table 5. NOTCH1, NOTCH3, HES1, and HEY1 primers were newly designed and GAPDH, JAG1, E6, and E7 primers were used as previously described [
7,
24,
25,
26]. After normalization, the ΔΔ
Ct method was used to compare relative expression for NOTCH1 pathway components and gel electrophoresis for HPV E6 and E7.
4.5. Immunohistochemical Study
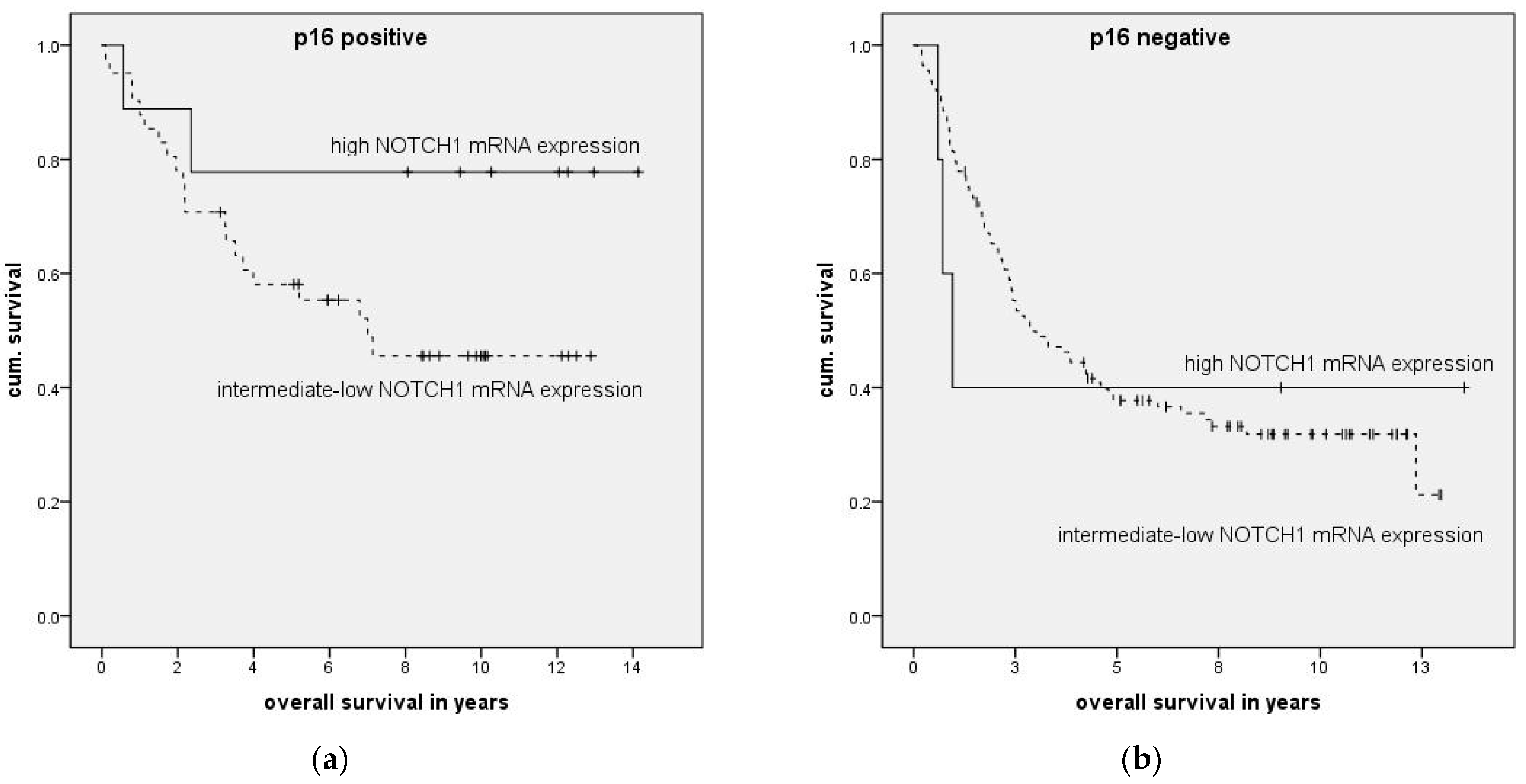
To identify carcinomas associated with HPV infection, p16 expression was analyzed in tumor tissue. From previously identified FFPE blocks, 1.5 µm sections (cut with Microm HM 355 S (International GmbH, Walldorf, Germany)) were placed on glass slides, dewaxed, and rehydrated. Microwave oven heating in citrate-buffered saline was used for antigen retrieval, as recommended by the manufacturer. After cooling, the slides were incubated with the antibody. A CINtec® Histology Kit (Roche Diagnostics GmbH, Mannheim, Germany) containing mouse monoclonal anti-p16INK4a (E6H4) at concentration 1 µg/ml was used according to the manufacturer’s protocol. Tissue with known expression of p16 was used as a positive control.
P16 expression level was described using a scoring system including staining intensity and percentage of stained tumor cells (
Table 6). HPV positivity was considered at 3 or more points.
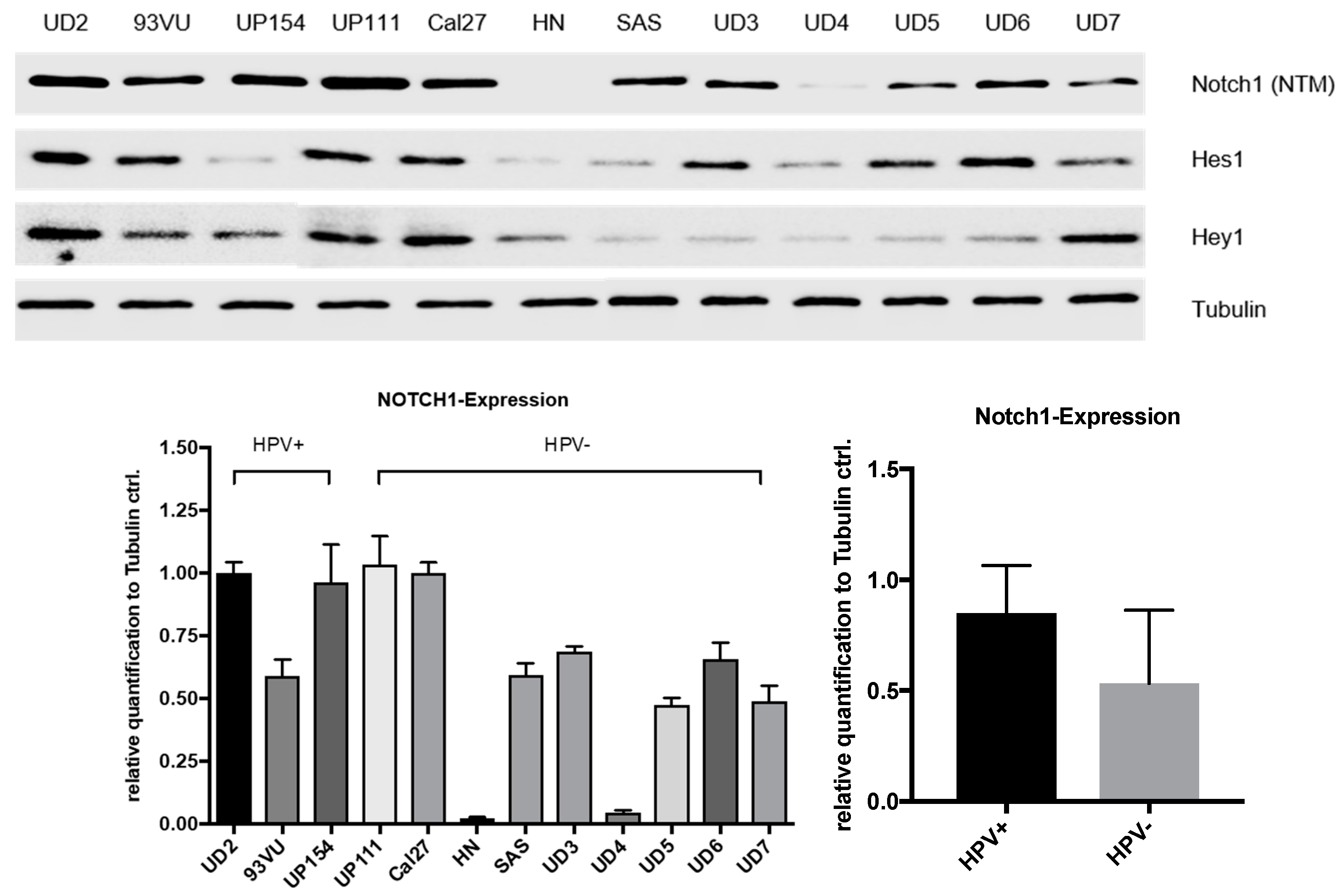
4.6. Cell Culture
The Cal27, HN, and UP-SCC-154 cell lines were obtained from DSMZ (Braunschweig, Germany), the UD-SCC-2-7 cell lines were obtained from the University of Düsseldorf (Department of Otorhinolaryngology, Düsseldorf, Germany), the 93VU cell line from VU University Medical Center Amsterdam (Department of Clinical Genetics, Amsterdam, Netherlands), and SAS from JCRB cell bank (Osaka, Japan). All cell lines have been STR profiled and were routinely prophylactically treated against mycoplasma infection. The cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Darmstadt, Germany) containing 10% fetal bovine serum (FBS) (Biochrom, Berlin, Germany), 2 mM glutamine, 100 µg/mL streptomycin, and 100 U/mL penicillin (Biochrom), maintained at 37 °C in an atmosphere of 5% CO2, and grown to 70–90% confluence.
4.7. Western Blot Analysis
For protein analysis, cells were grown to 70% confluence in 10 cm tissue culture dishes. Cells were washed with ice-cold 1× DPBS and lysed with 500 μL of cell lysis buffer. The buffer contained 1× Cell Lysis Buffer (Cell Signaling, Danvers, MA, USA), 1 mM PMSF (Carl Roth, Karlsruhe, Germany), and 1× Protease Inhibitory Cocktail (Cell Signaling). The lysis buffer (10×) (Cell Signaling) included 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 µg/mL leupeptin. Next, the cells were scraped off the culture dishes, pipetted into 1.5 mL microtubes, incubated on ice, and centrifuged at 4 °C and 10,000 rpm for 15 min to isolate the soluble protein fraction. The clarified lysate was frozen at −20 °C until use in the Bradford assay. The Bradford assay was used to verify that equal amounts were loaded per lane on an SDS-PAGE.
Equal protein concentrations (15 μg) were separated for 3 h at 120 V using an SDS-PAGE (Blotting System Mini-PROTEAN® Tetra System and PowerPacTM HC from Bio-Rad Laboratories, Munich, Germany) in a Tris-glycine running buffer. The densities of the running gels ranged from 7.5% to 12.5%, and the stacking gels possessed a density of 5%. The proteins were then transferred to a polyvinylidene fluoride (PVDF) membrane (Merck Millipore, Darmstadt, Germany) using a Trans-Blot® SD Semi Dry Transfer Cell (Bio-Rad Laboratories, Munich, Germany) at 225 mA for 80 min. A solution containing 5% nonfat dry milk in 1× TBS and 0.1% Tween-20 was used to block unspecific binding sites. The membranes were then incubated with the primary antibodies against NOTCH1 (NTM), HES1, HEY1, and Tubulin (all from Cell Signaling Technologies, Danvers, MA, USA) in 1× TBS + 0.1% Tween-20 for 12 h at 4 °C, washed, and incubated with an HRP-linked secondary antibody (Cell Signaling technologies) in 5% nonfat dry milk in 1× TBS and 0.1% Tween-20 for 1 h at room temperature. Next, the membranes were washed and incubated in Thermo Scientific™ Pierce™ ECL Western Blotting Substrate (Fisher Scientific, Waltham, MA, USA) for 1 minute. Immunoreactivity was visualized by ChemiDoc XRS+ with Image LabTM Software (Bio-Rad Laboratories, Munich, Germany). Protein expression was quantified with scanning densitometry and values normalized to a tubulin control.
4.8. Statistical Analysis
All statistical tests were two-sided and significance was determined at a level of 5%. For comparison of mRNA expression in normal tissue versus tumor tissue and p16-positive and p16-negative tissue, Mann–Whitney U Test or Kruskal–Wallis Test was used. Correlation was calculated according to Spearman Rho. Expression in Western blots was compared with t-test.
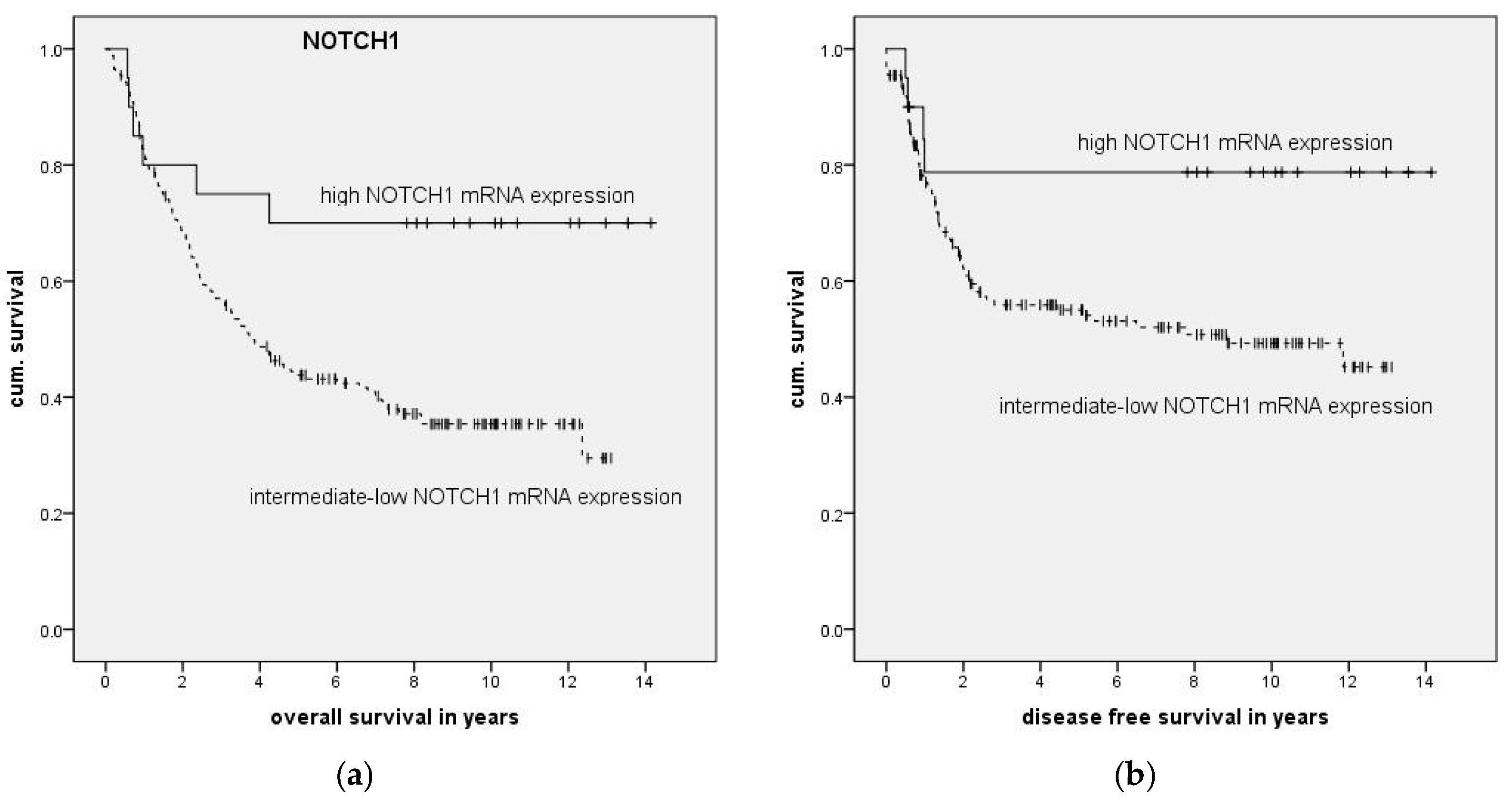
To examine the impact of mRNA expression on clinical parameters we categorized patients into high and low expression groups and compared these to the remaining patients. High and low mRNA expression was defined as mean relative mRNA expression plus or minus one standard deviation.
Association of clinical parameters and high NOTCH1 expression was compared with Fisher’s exact test with Bonferroni correction applied. The impact of expression levels on survival was analyzed with Kaplan–Meier curves, and significance was calculated using log-rank testing. To examine the association of expression levels with clinical data, multivariate forward stepwise Cox regression was performed. Statistical calculations were done in SPSS version 23 (IBM, Ehningen, Germany) or GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA).