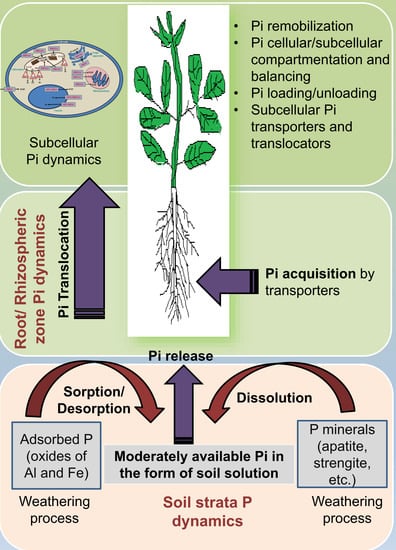
Cellular and Subcellular Phosphate Transport Machinery in Plants
Abstract
:1. Introduction
2. Phosphate Transporters and Their Role in Pi Acquisition, Translocation, and Remobilization in Various Organs
3. Subcellular Pi Transport and Balancing
3.1. Vacuole: The Pi Storehouse
3.2. Chloroplast
3.3. Mitochondria
3.4. Cellular Movement of Pi Transporters Involves the ER
3.5. Golgi Bodies
4. Conclusions and Future Research Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CK2β3 | Casein kinase 2β3 |
| ESCRT | Endosomal complex required for transport |
| GPT | Glucose-6-phosphate/phosphate translocator |
| MPT | Membrane-localized phosphate transporter |
| NLA | Nitrogen limitation adaptation |
| SNF | Sucrose non-fermenting protein |
| SPX-MFS | SYG1/PHO81/XPR major facilitator superfamily |
| P | Phosphorus |
| PHO | Phosphate transporter |
| PHF | Phosphate transporter traffic facilitator |
| PHT | High-affinity phosphate transporter |
| Pi | Phosphate |
| PPT | Phosphoenolpyruvate/phosphate translocator |
| pPT | Plastidic phosphate translocator |
| XPT | Xylulose-5-phosphate/phosphate translocator |
| VPT | Vacuolar phosphate transporter |
References
- Liu, J.; Yang, L.; Luan, M.; Wang, Y.; Zhang, C.; Zhang, B.; Shi, J.; Zhao, F.G.; Lan, W.; Luan, S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E6571–E6578. [Google Scholar] [CrossRef] [PubMed]
- Kleinert, A.; Thuynsma, R.; Magadlela, A.; Benedito, V.A.; Valentine, A.J. Metabolism and transport of carbon in legume nodules under phosphorus deficiency. In Legume Nitrogen Fixation in Soils with Low Phosphorus Availability; Sulieman, S., Tran, L.S.P., Eds.; Springer: Cham, The Switzerland, 2017; pp. 77–95. [Google Scholar]
- Shen, J.; Yuan, L.; Zhang, J.; Li, H.; Bai, Z.; Chen, X.; Zhang, W.; Zhang, F. Phosphorus dynamics: From soil to plant. Plant Physiol. 2011, 156, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, M.; El-Sayed, M.; Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Burritt, D.J.; Tran, L.P. Metabolomics and transcriptomics in legumes under phosphate deficiency in relation to nitrogen fixation by root nodules. Front. Plant Sci. 2018. [Google Scholar] [CrossRef]
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Chen, A.; Sun, S.; Xu, G. Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol. Plant 2016, 9, 396–416. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Y.; Huang, T.K.; Kuo, H.F.; Chiou, T. Role of vacuoles in phosphorus storage and remobilization. J. Exp. Bot. 2017, 68, 3045–3055. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.; Boisson, A.M.; Gout, E.; Bligny, R.; Douce, R.; Aubert, S. Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: An in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol. 2009, 151, 1646–1657. [Google Scholar] [CrossRef] [PubMed]
- Lapis-Gaza, H.R.; Jost, R.; Finnegan, P.M. Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 2014, 14, 334. [Google Scholar] [CrossRef] [PubMed]
- Kisko, M.; Shukla, V.; Kaur, M.; Bouain, N.; Chaiwong, N.; Lacombe, B.; Pandey, A.K.; Rouached, H. Phosphorus transport in Arabidopsis and wheat: Emerging strategies to improve P pool in seeds. Agriculture 2018, 8, 27. [Google Scholar] [CrossRef]
- Schachtman, D.P.; Reid, R.J.; Ayling, S.M. Phosphorus uptake by plants: From soil to cell. Plant Physiol. 1998, 116, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta 2002, 216, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Sakano, K.; Yazaki, Y.; Mimura, T. Cytoplasmic acidification induced by inorganic phosphate uptake in suspension cultured Catharanthus roseus cells—Measurement with fluorescent pH indicator and P-31 nuclear-magnetic-resonance. Plant Physiol. 1992, 99, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, C.I.; Novacky, A.J. Extra- and intracellular pH and membrane potential changes induced by K+, Cl‒, H2PO4‒, and NO3‒ uptake and fusicoccin in root hairs of Limnobium stoloniferum. Plant Physiol. 1990, 94, 1561–1567. [Google Scholar] [CrossRef] [PubMed]
- Versaw, W.K.; Garcia, L.R. Intracellular transport and compartmentation of phosphate in plants. Curr. Opin. Plant Biol. 2017, 39, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Lambers, H.; Finnegan, P.M.; Jost, R.; Plaxton, W.C.; Shane, M.W.; Stitt, M. Phosphorus nutrition in proteaceae and beyond. Nat. Plants 2015, 1, 15109. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant. 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Igamberdiev, A.U.; Kleczkowski, L.A. Membrane potential, adenylate levels and Mg2+ are interconnected via adenylate kinase equilibrium in plant cells. Biochim. Biophys. Acta 2003, 1607, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yue, W.; Ying, Y.; Wang, S.; Secco, D.; Liu, Y.; Whelan, J.; Tyerman, S.D.; Shou, H. Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 2015, 169, 2822–2831. [Google Scholar] [CrossRef] [PubMed]
- Chandy, G.; Grabe, M.; Moore, H.P.; Machen, T.E. Proton leak and CFTR in regulation of Golgi pH in respiratory epithelial cells. Am. J. Physiol. Cell Physiol. 2001, 281, C908–C921. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Peters, J.; Berkowitz, G.A. Surface charge-mediated effects of Mg2+ on K+ flux across the chloroplast envelope are associated with regulation of stromal pH and photosynthesis. Plant Physiol. 1991, 97, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Kramer, D.M.; Cruz, J.A.; Kanazawa, A. Balancing the central roles of the thylakoid proton gradient. Trends Plant Sci. 2003, 8, 27–32. [Google Scholar] [CrossRef]
- Sun, T.; Li, M.; Shao, Y.; Yu, L.; Ma, F. Comprehensive genomic identification and expression analysis of the phosphate transporter (PHT) gene family in apple. Front. Plant Sci. 2017, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Ragothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs, and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low-and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, A.S.; Varadarajan, D.K.; Mukatira, U.T.; D’Urzo, M.P.; Damsz, B.; Raghothama, K.G. Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 2002, 130, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014, 164, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Yuan, J.; Chang, X.; Yang, M.; Zhang, L.; Lu, K.; Lian, X. The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 2015, 10, e0126186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sun, Y.; Pei, W.; Jain, A.; Sun, R.; Cao, Y.; Wu, X.; Jiang, T.; Zhang, L.; Fan, X.; et al. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 2015, 82, 556–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.Y.; Gronlund, M.; Jakobsen, I.; Grotemeyer, M.S.; Rentsch, D.; Miyao, A.; Hirochika, H.; Kumar, C.S.; Sundaresan, V.; Salamin, N.; et al. Non redundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the PHOSPHATE TRANSPORTER1 gene family. Plant Cell 2012, 24, 4236–4251. [Google Scholar] [CrossRef] [PubMed]
- Arpat, A.B.; Magliano, P.; Wege, S.; Rouached, H.; Stefanovic, A.; Poirier, Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012, 71, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, A.; Ribot, C.; Rouached, H.; Wang, Y.; Chong, J.; Belbahri, L.; Delessert, S.; Poirier, Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007, 50, 982–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, G.A.; Bouraine, S.; Wege, S.; Li, Y.Y.; de Carbonnel, M.; Berthomieu, P.; Poirier, Y.; Rouached, H. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 2014, 65, 871–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luan, M.; Tang, R.J.; Tang, Y.; Tian, W.; Hou, C.; Zhao, F.; Lan, W.; Luan, S. Transport and homeostasis of potassium and phosphate: Limiting factors for sustainable crop production. J. Exp. Bot. 2016, 68, 3091–3105. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.; Sun, S.; Zhao, J.; Fan, X.; Xin, W.; Guo, Q.; Yu, L.; Shen, Q.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.P.; Nagarajan, K.V.; Raghothama, K.G. Arabidopsis Pht1;5 plays an integral role in phosphate homeostasis. Plant Signal. Behav. 2011, 6, 1676–1678. [Google Scholar] [CrossRef] [PubMed]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Ayadi, A.; David, P.; Arrighi, J.F.; Chiarenza, S.; Thibaud, M.C.; Nussaume, L.; Marin, E. Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 2015, 167, 1511–1526. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, D.; Rezzonico, E.; MacDonald-Comber Petetot, J.; Somerville, C.; Poirier, Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Thomas, S.; Somerville, C.; Schiefelbein, J. Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 1991, 97, 1087–1093. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Baumann, A.; Poirier, Y. Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 2010, 152, 1693–1704. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wu, X.N.; Zhou, H.M.; Wang, D.F.; Jiang, T.T.; Sun, Y.F.; Cao, Y.; Pei, W.X.; Sun, S.B.; Xu, G.H. Overexpression of rice phosphate transporter gene OsPT6 enhances phosphate uptake and accumulation in transgenic rice plants. Plant Soil 2014, 384, 259–270. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Zhang, X.; Fan, H.; Gu, M.; Qu, H.; Xu, G. Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 2015, 230, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Ren, H.; Gu, M.; Zhao, J.; Sun, S.; Zhang, X.; Chen, J.; Wu, P.; Xu, G. The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant. Physiol. 2011, 156, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Huang, T.K.; Yang, S.Y.; Hong, Y.T.; Huang, S.M.; Wang, F.N.; Chiang, S.F.; Tsai, S.Y.; Lu, W.C.; Chiou, T.J. Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 2016, 7, 110965. [Google Scholar] [CrossRef] [PubMed]
- Gilliham, M.; Athman, A.; Tyerman, S.D.; Conn, S.J. Cell-specific compartmentation of mineral nutrients is an essential mechanism for optimal plant productivity—Another role for TPC1? Plant Sign Behav. 2011, 6, 1656–1661. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.; Gilliham, M. Comparative physiology of elemental distributions in plants. Ann. Bot. 2010, 105, 1081–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Versaw, W.K.; Harrison, M.J. A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 2002, 14, 1751–1766. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Irigoyen, S.; Fowler, T.B.; Versaw, W.K. Differential expression and phylogenetic analysis suggest specialization of plastid-localized members of the PHT4 phosphate transporter family for photosynthetic and heterotrophic tissues. Plant Signal. Behav. 2008, 3, 784–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irigoyen, S.; Karlsson, P.M.; Kuruvilla, J.; Spetea, C.; Versaw, W.K. The sink-specific plastidic phosphate transporter PHT4;2 influences starch accumulation and leaf size in Arabidopsis. Plant Physiol. 2011, 157, 1765–1777. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Miao, Q.; Sun, D.; Yang, G.; Wu, C.; Huang, J.; Zheng, C. The mitochondrial phosphate transporters modulate plant response to salt stress via affecting ATP and gibberellin metabolism in Arabidopsis thaliana. PLoS ONE 2012, 7, e43530. [Google Scholar] [CrossRef] [PubMed]
- Hamel, P.; Saint-Georges, Y.; de Pinto, B.; Lachacinski, N.; Altamura, N.; Dujardin, G. Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol. Microbiol. 2004, 51, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Poirier, Y.; Bucher, M. Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 2002, 1, e0024. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Chang, X.J.; Ye, Y.; Xie, W.B.; Wu, P.; Lian, X.M. Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 2011, 4, 1105–1122. [Google Scholar] [CrossRef] [PubMed]
- Hassler, S.; Lemke, L.; Jung, B.; Möhlmann, T.; Krüger, F.; Schumacher, K.; Espen, L.; Martinoia, E.; Neuhaus, H.E. Lack of the Golgi phosphate transporter PHT4;6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation in Arabidopsis. Plant J. 2012, 72, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Hassler, S.; Jung, B.; Lemke, L.; Novak, O.; Strnad, M.; Martinoia, E.; Neuhaus, H.E. Function of the Golgi located phosphate transporter PHT4;6 is critical for senescence associated processes in Arabidopsis. J. Exp. Bot. 2016, 67, 4671–4684. [Google Scholar] [CrossRef] [PubMed]
- Flügge, U.I.; Hausler, R.E.; Ludewig, F.; Gierth, M. The role of transporters in supplying energy to plant plastids. J. Exp. Bot. 2011, 62, 2381–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knappe, S.; Lottgert, T.; Schneider, A.; Voll, L.; Flugge, U.I.; Fischer, K. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 2003, 36, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, H.E.; Thom, E.; Mohlmann, T.; Steup, M.; Kampfenkel, K. Characterization of a novel eukaryotic ATP/ADP translocator located in the plastid envelope of Arabidopsis thaliana L. Plant J. 1997, 11, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Eom, J.S.; Voll, L.M.; Prasch, C.M.; Park, Y.L.; Hahn, T.R.; Ha, S.H.; An, G.; Jeon, J.S. Analysis of a triose phosphate/phosphate translocator-deficient mutant reveals a limited capacity for starch synthesis in rice leaves. Mol. Plant 2014, 7, 1705–1708. [Google Scholar] [CrossRef] [PubMed]
- Bornke, F.; Sonnewald, S. Biosynthesis and metabolism of starch and sugars. In Plant Metabolism and Biotechnology; Ashihara, H., Crozier, A., Komamine, A., Eds.; John Wiley & Sons: Chichester, UK, 2011; pp. 1–25. ISBN 978-0-470-74703-2. [Google Scholar]
- Guo, B.; Jin, Y.; Wussler, C.; Blancaflor, E.B.; Motes, C.M.; Versaw, W.K. Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 2008, 177, 889–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, P.M.; Herdean, A.; Adolfsson, L.; Beebo, A.; Nziengui, H.; Irigoyen, S.; Unnep, R.; Zsiros, O.; Nagy, G.; Garab, G.; et al. The Arabidopsis thylakoid transporter PHT4;1 influences phosphate availability for ATP synthesis and plant growth. Plant J. 2015, 84, 99–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haferkamp, I.; Hackstein, J.H.; Voncken, F.G.; Schmit, G.; Tjaden, J. Functional integration of mitochondrial and hydrogenosomal ADP/ATP carriers in the Escherichia coli membrane reveals different biochemical characteristics for plants, mammals and anaerobic chytrids. Eur. J. Biochem. 2002, 269, 3172–3181. [Google Scholar] [CrossRef] [PubMed]
- Laloi, M. Plant mitochondrial carriers: An overview. Cell. Mol. Life Sci. 1999, 56, 918–944. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Lytovchenko, A.; Morgan, M.; Nunes-Nesi, A.; Taylor, N.L.; Baxter, C.J.; Eickmeier, I.; Fernie, A.R. Mitochondrial uncoupling protein is required for efficient photosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19587–19592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, F.; Wan, X.; Zhu, W.; Sun, D.; Zheng, C.; Liu, P.; Huang, J. Overexpression of mitochondrial phosphate transporter 3 severely hampers plant development through regulating mitochondrial function in Arabidopsis. PLoS ONE 2015, 10, e0129717. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high affinity phosphate transporter in Arabidopsis. Plant Cell 2005, 17, 3500–3512. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Guo, M.; Cai, L.; Hu, H.; Li, C.; Liu, Y.; Wu, Z.; Mao, C.; Yi, K.; Wu, P.; et al. Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol. Biol. 2015, 87, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Z.; Ren, H.; Shen, C.; Li, Y.; Ling, H.Q.; Wu, C.; Lian, X.; Wu, P. OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 2010, 62, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, Y.; Ni, J.; Wang, Y.; Bai, Y.; Shi, J.; Gan, J.; Wu, Z.; Wu, P. OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 2011, 157, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Wang, F.; Yang, J.; Gao, M.; Li, C.; Liu, Y.; Liu, Y.; Yamaji, N.; Ma, J.F.; et al. The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 2015, 27, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Cardona-López, X.; Cuyas, L.; Marin, E.; Rajulu, C.; Irigoyen, M.L.; Gil, E.; Puga, M.I.; Bligny, R.; Nussaume, L.; Geldner, N.; et al. ESCRT-III-associated protein ALIX mediates high-affinity phosphate transporter trafficking to maintain phosphate homeostasis in Arabidopsis. Plant Cell 2015, 27, 2560–2581. [Google Scholar] [CrossRef] [PubMed]
- Bayle, V.; Arrighi, J.F.; Creff, A.; Nespoulous, C.; Vialaret, J.; Rossignol, M.; Gonzalez, E.; Paz-Ares, J.; Nussaume, L. Arabidopsis thaliana high affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 2011, 23, 1523–1535. [Google Scholar] [CrossRef] [PubMed]
- Driouich, A.; Follet-Gueye, M.L.; Bernard, S.; Kousar, S.; Chevalier, L.; Vicré-Gibouin, M.; Lerouxel, O. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 2012, 3, 79. [Google Scholar] [CrossRef] [PubMed]
- Cubero, B.; Nakagawa, Y.; Jiang, X.Y.; Miura, K.J.; Li, F.; Raghothama, K.G.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant 2009, 2, 535–552. [Google Scholar] [CrossRef] [PubMed]


| Subcellular Organelle | pH | Δψ (Membrane Potential) | References | |
|---|---|---|---|---|
| Mitochondrion | 8.1 | From −90 to −120 mV | [17,18] | |
| Vacuole | 5.2 | +31 mV | [17,19] | |
| Golgi body | 6.3 | Not known in plants | [17,20] | |
| Plastid (non-photosynthetic) | 7.3 | −144 mV | [21] | |
| Photosynthetic plastid | Thylakoid lumen | 5.8–6.5 | +30 mV | [22] |
| Chloroplast stroma | 8.0 | −123 mV | [17,21] | |
| Cytosol | 7.3 | −172 mV | [17] | |
| Transporter(s) | Function | References |
|---|---|---|
| Root Uptake | ||
| AtPHT1;1, AtPHT1;2, AtPHT1;3, AtPHT1;4 | Involved in Pi uptake | [9,24,25,26] |
| OsPHT1;1, OsPHT1;2, OsPHT1;4, OsPHT1;6, OsPHT1;9, OsPHT1;10, OsPHT1;11, OsPHT1;13 | Involved in Pi uptake/translocation. OsPHT1;11 and OsPHT1;13 play roles in Pi uptake in symbiotic association with arbuscular mycorrhizal fungi. | [27,28,29,30] |
| Root-to-Shoot Translocation | ||
| AtPHT1;8, AtPHT1;9, AtPHO1, AtPHO1;H1, AtPHO1;H3 | Translocation of Pi from roots to shoots. AtPHO1;H3 is involved in the suppression of root-to-shoot Pi transport under Zn-deficient conditions. | [9,31,32,33] |
| OsPHT1;2, OsPHT1;4, OsPHT1;6, OsPHT1;8, OsPHO1;2 | Translocation of Pi from roots to shoots. | [34,35] |
| Pi Redistribution and Remobilization | ||
| AtPHT1;1, AtPHT1;5, AtPHT1;9 | AtPHT1;5 plays a role in Pi translocation from source to sink organs. Pi redistribution across the vegetative organs. | [24,36,37,38] |
| OsPHT1;4, OsPHT1;6, OsPHT1;8 | OsPHT1;4 is involved in the remobilization of Pi from flag leaves to the panicles. OsPHT1;6 and OsPHT1;8 help in Pi remobilization from senescing leaves to young leaves and rice grains. | [3] |
| Transporter(s) | Function | References |
|---|---|---|
| Vacuole | ||
| OsSPXMFS1, AtVPT1/AtPHT5;1 | Import | [1,19,48] |
| OsSPXMFS1 OsSPXMFS3 | Import Export and symport | [19,48] |
| Chloroplast | ||
| AtPHT2;1, AtPHT4;1, ANTR1 (leaf chloroplast) AtPHT4;2 (root plastid) AtPHT4;3 (shoot plastid) AtPHT4;4 (leaf chloroplast) AtPHT4;5 (shoot plastid) | Import and symport | [51,52,53] |
| Mitochondrion | ||
| AtPHT3;1, AtPHT3;2, AtPHT3;3 | Import and symport | [54,55,56] |
| OsPT15 (located on peroxisome) OsPT16 (located on endoplasmic reticulum) OsPT17 (located on peroxisome) OsPT18 (located on peroxisome) OsPT19 (located on peroxisome) OsPT20 (located on plasma membrane) | Import and symport | [57] |
| Golgi Body | ||
| AtPHT4;6 | Export | [58,59] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, S.; Upadhyay, M.K.; Srivastava, A.K.; Abdelrahman, M.; Suprasanna, P.; Tran, L.-S.P. Cellular and Subcellular Phosphate Transport Machinery in Plants. Int. J. Mol. Sci. 2018, 19, 1914. https://doi.org/10.3390/ijms19071914
Srivastava S, Upadhyay MK, Srivastava AK, Abdelrahman M, Suprasanna P, Tran L-SP. Cellular and Subcellular Phosphate Transport Machinery in Plants. International Journal of Molecular Sciences. 2018; 19(7):1914. https://doi.org/10.3390/ijms19071914
Chicago/Turabian StyleSrivastava, Sudhakar, Munish Kumar Upadhyay, Ashish Kumar Srivastava, Mostafa Abdelrahman, Penna Suprasanna, and Lam-Son Phan Tran. 2018. "Cellular and Subcellular Phosphate Transport Machinery in Plants" International Journal of Molecular Sciences 19, no. 7: 1914. https://doi.org/10.3390/ijms19071914