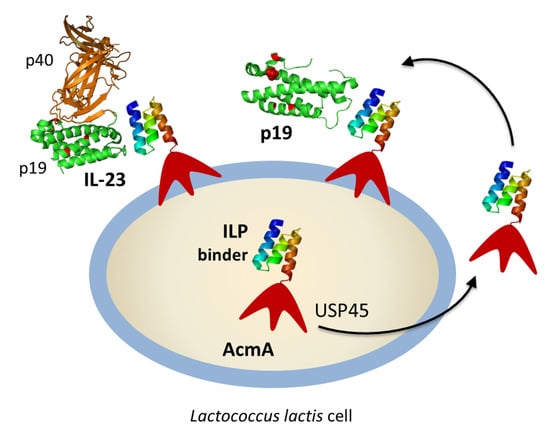
p19-Targeting ILP Protein Blockers of IL-23/Th-17 Pro-Inflammatory Axis Displayed on Engineered Bacteria of Food Origin
Abstract
:1. Introduction
2. Results
2.1. Molecular Assembly of Recombinant p19 Protein
2.2. Expression of ILP-Fusion Proteins in L. lactis
2.3. Display of ILP-Fusion Proteins on the Surface of L. lactis
2.4. Binding of Recombinant p19 by ILP-Displaying L. lactis
2.5. Removal of IL-23 by ILP-Displaying L. lactis
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Media and Growth Conditions
4.2. DNA Manipulation and Plasmid Construction
4.3. Production of p19-TRX Fusion Protein
4.4. p19 Binding Assay
4.5. Expression of Cytokine Binding Fusion Proteins in L. lactis
4.6. SDS PAGE and Western Blot
4.7. Flow Cytometry
4.8. IL-23 Binding Assay
4.9. Statistics
4.10. Modeling of ILP-p19 and Interactions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Girolomoni, G.; Strohal, R.; Puig, L.; Bachelez, H.; Barker, J.; Boehncke, W.H.; Prinz, J.C. The role of IL-23 and the IL-23/TH 17 immune axis in the pathogenesis and treatment of psoriasis. J. Eur. Acad Dermatol. Venereol. 2017, 31, 1616–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razawy, W.; van Driel, M.; Lubberts, E. The role of IL-23 receptor signaling in inflammation-mediated erosive autoimmune arthritis and bone remodeling. Eur. J. Immunol. 2018, 48, 220–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, C.; Zhang, H. The Role of Proinflammatory Pathways in the Pathogenesis of Colitis-Associated Colorectal Cancer. Mediat. Inflamm. 2017, 2017, 5126048. [Google Scholar] [CrossRef] [PubMed]
- Sarra, M.; Pallone, F.; Macdonald, T.T.; Monteleone, G. IL-23/IL-17 axis in IBD. Inflamm. Bowel. Dis. 2010, 16, 1808–1813. [Google Scholar] [CrossRef] [PubMed]
- Siakavellas, S.I.; Bamias, G. Role of the IL-23/IL-17 axis in Crohn’s disease. Discov. Med. 2012, 14, 253–262. [Google Scholar] [PubMed]
- Floss, D.M.; Schroder, J.; Franke, M.; Scheller, J. Insights into IL-23 biology: From structure to function. Cytokine Growth Factor Rev. 2015, 26, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Gaffen, S.L.; Jain, R.; Garg, A.V.; Cua, D.J. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 2014, 14, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Bilal, J.; Berlinberg, A.; Bhattacharjee, S.; Trost, J.; Riaz, I.B.; Kurtzman, D.J.B. A Systematic Review and Meta-Analysis of the Efficacy and Safety of the Interleukin (IL)-12/23 and IL-17 Inhibitors Ustekinumab, Secukinumab, Ixekizumab, Brodalumab, Guselkumab, and Tildrakizumab for the Treatment of Moderate to Severe Plaque Psoriasis. J. Dermatol. Treat. 2018, 1–37. [Google Scholar] [CrossRef] [PubMed]
- Fotiadou, C.; Lazaridou, E.; Sotiriou, E.; Ioannides, D. Targeting IL-23 in psoriasis: Current perspectives. Psoriasis (Auckl) 2018, 8, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, A.; Spinelli, S.; Boutton, C.; Saunders, M.; Blachetot, C.; de Haard, H.; Denecker, G.; Van Roy, M.; Cambillau, C.; Rommelaere, H. Neutralization of Human Interleukin 23 by Multivalent Nanobodies Explained by the Structure of Cytokine-Nanobody Complex. Front. Immunol. 2017, 8, 884. [Google Scholar] [CrossRef] [PubMed]
- Ramamurthy, V.; Krystek, S.R., Jr.; Bush, A.; Wei, A.; Emanuel, S.L.; Das Gupta, R.; Janjua, A.; Cheng, L.; Murdock, M.; Abramczyk, B.; et al. Structures of adnectin/protein complexes reveal an expanded binding footprint. Structure 2012, 20, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Desmet, J.; Verstraete, K.; Bloch, Y.; Lorent, E.; Wen, Y.; Devreese, B.; Vandenbroucke, K.; Loverix, S.; Hettmann, T.; Deroo, S.; et al. Structural basis of IL-23 antagonism by an Alphabody protein scaffold. Nat. Commun. 2014, 5, 5237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuchar, M.; Vankova, L.; Petrokova, H.; Cerny, J.; Osicka, R.; Pelak, O.; Sipova, H.; Schneider, B.; Homola, J.; Sebo, P.; et al. Human interleukin-23 receptor antagonists derived from an albumin-binding domain scaffold inhibit IL-23-dependent ex vivo expansion of IL-17-producing T-cells. Proteins 2014, 82, 975–989. [Google Scholar] [CrossRef] [PubMed]
- Krizova, L.; Kuchar, M.; Petrokova, H.; Osicka, R.; Hlavnickova, M.; Pelak, O.; Cerny, J.; Kalina, T.; Maly, P. p19-targeted ABD-derived protein variants inhibit IL-23 binding and exert suppressive control over IL-23-stimulated expansion of primary human IL-17+ T-cells. Autoimmunity 2017, 50, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Worledge, K.L.; Godiska, R.; Barrett, T.A.; Kink, J.A. Oral administration of avian tumor necrosis factor antibodies effectively treats experimental colitis in rats. Dig. Dis. Sci. 2000, 45, 2298–2305. [Google Scholar] [CrossRef] [PubMed]
- Pawar, V.K.; Meher, J.G.; Singh, Y.; Chaurasia, M.; Surendar Reddy, B.; Chourasia, M.K. Targeting of gastrointestinal tract for amended delivery of protein/peptide therapeutics: Strategies and industrial perspectives. J. Control. Release 2014, 196, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Braat, H.; Rottiers, P.; Hommes, D.W.; Huyghebaert, N.; Remaut, E.; Remon, J.P.; van Deventer, S.J.; Neirynck, S.; Peppelenbosch, M.P.; Steidler, L. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 2006, 4, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; de Vos, W.M. Functional genomics of lactic acid bacteria: From food to health. Microb. Cell Factor 2014, 13. [Google Scholar] [CrossRef] [PubMed]
- Sanders, M.E. Probiotics: Definition, sources, selection, and uses. Clin. Infect. Dis. 2008, 46 (Suppl. 2), S58–S61, discussion S144–S51. [Google Scholar] [CrossRef] [PubMed]
- Siezen, R.J.; Bayjanov, J.R.; Felis, G.E.; van der Sijde, M.R.; Starrenburg, M.; Molenaar, D.; Wels, M.; van Hijum, S.A.; van Hylckama Vlieg, J.E. Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb. Biotechnol. 2011, 4, 383–402. [Google Scholar] [CrossRef] [PubMed]
- Ballal, S.A.; Veiga, P.; Fenn, K.; Michaud, M.; Kim, J.H.; Gallini, C.A.; Glickman, J.N.; Quere, G.; Garault, P.; Beal, C.; et al. Host lysozyme-mediated lysis of Lactococcus lactis facilitates delivery of colitis-attenuating superoxide dismutase to inflamed colons. Proc. Natl. Acad. Sci. USA 2015, 112, 7803–7808. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, M.; Nemoto, M.; Nakata, T.; Kondo, S.; Takahashi, H.; Kimura, B.; Kuda, T. Anti-inflammatory properties of fermented soy milk with Lactococcus lactis subsp. lactis S-SU2 in murine macrophage RAW264.7 cells and DSS-induced IBD model mice. Int. Immunopharmacol. 2015, 26, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Drouault, S.; Corthier, G.; Ehrlich, S.D.; Renault, P. Survival, physiology, and lysis of Lactococcus lactis in the digestive tract. Appl. Environ. Microbiol. 1999, 65, 4881–4886. [Google Scholar] [PubMed]
- Berlec, A.; Ravnikar, M.; Strukelj, B. Lactic acid bacteria as oral delivery systems for biomolecules. Pharmazie 2012, 67, 891–898. [Google Scholar] [PubMed]
- Bermudez-Humaran, L.G.; Aubry, C.; Motta, J.P.; Deraison, C.; Steidler, L.; Vergnolle, N.; Chatel, J.M.; Langella, P. Engineering lactococci and lactobacilli for human health. Curr. Opin. Microbiol. 2013, 16, 278–283. [Google Scholar] [CrossRef] [PubMed]
- Song, A.A.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcus lactis: From food to factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, K.; de Haard, H.; Beirnaert, E.; Dreier, T.; Lauwereys, M.; Huyck, L.; Van Huysse, J.; Demetter, P.; Steidler, L.; Remaut, E.; et al. Orally administered L. lactis secreting an anti-TNF Nanobody demonstrate efficacy in chronic colitis. Mucosal. Immunol. 2010, 3, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Perse, M.; Ravnikar, M.; Lunder, M.; Erman, A.; Cerar, A.; Strukelj, B. Dextran sulphate sodium colitis in C57BL/6J mice is alleviated by Lactococcus lactis and worsened by the neutralization of Tumor necrosis Factor alpha. Int. Immunopharmacol. 2017, 43, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, K.; Hans, W.; Van Huysse, J.; Neirynck, S.; Demetter, P.; Remaut, E.; Rottiers, P.; Steidler, L. Active delivery of trefoil factors by genetically modified Lactococcus lactis prevents and heals acute colitis in mice. Gastroenterology 2004, 127, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, H.J.; Wiepjes, M.; Motta, J.P.; Schulz, J.D.; Jury, J.; Natividad, J.M.; Pinto-Sanchez, I.; Sinclair, D.; Rousset, P.; Martin-Rosique, R.; et al. Novel role of the serine protease inhibitor elafin in gluten-related disorders. Am. J. Gastroenterol. 2014, 109, 748–756. [Google Scholar] [CrossRef] [PubMed]
- Steidler, L.; Hans, W.; Schotte, L.; Neirynck, S.; Obermeier, F.; Falk, W.; Fiers, W.; Remaut, E. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 2000, 289, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Ravnikar, M.; Strukelj, B.; Obermajer, N.; Lunder, M.; Berlec, A. Engineered lactic acid bacterium Lactococcus lactis capable of binding antibodies and tumor necrosis factor alpha. Appl. Environ. Microbiol. 2010, 76, 6928–6932. [Google Scholar] [CrossRef] [PubMed]
- Skrlec, K.; Pucer Janez, A.; Rogelj, B.; Strukelj, B.; Berlec, A. Evasin-displaying lactic acid bacteria bind different chemokines and neutralize CXCL8 production in Caco-2 cells. Microb. Biotechnol. 2017, 10, 1732–1743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michon, C.; Langella, P.; Eijsink, V.G.; Mathiesen, G.; Chatel, J.M. Display of recombinant proteins at the surface of lactic acid bacteria: Strategies and applications. Microb. Cell Factor. 2016, 15, 70. [Google Scholar] [CrossRef] [PubMed]
- Dieye, Y.; Usai, S.; Clier, F.; Gruss, A.; Piard, J.C. Design of a protein-targeting system for lactic acid bacteria. J. Bacteriol. 2001, 183, 4157–4166. [Google Scholar] [CrossRef] [PubMed]
- Buist, G.; Kok, J.; Leenhouts, K.J.; Dabrowska, M.; Venema, G.; Haandrikman, A.J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 1995, 177, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Steen, A.; Buist, G.; Horsburgh, G.J.; Venema, G.; Kuipers, O.P.; Foster, S.J.; Kok, J. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J. 2005, 272, 2854–2868. [Google Scholar] [CrossRef] [PubMed]
- Kosler, S.; Strukelj, B.; Berlec, A. Lactic Acid Bacteria with Concomitant IL-17, IL-23 and TNFalpha-Binding Ability for the Treatment of Inflammatory Bowel Disease. Curr. Pharm. Biotechnol. 2017, 18, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Bloch, Y.; Bouchareychas, L.; Merceron, R.; Skladanowska, K.; Van den Bossche, L.; Detry, S.; Govindarajan, S.; Elewaut, D.; Haerynck, F.; Dullaers, M.; et al. Structural Activation of Pro-inflammatory Human Cytokine IL-23 by Cognate IL-23 Receptor Enables Recruitment of the Shared Receptor IL-12Rbeta1. Immunity 2018, 48, 45–58.e6. [Google Scholar] [CrossRef] [PubMed]
- Holo, H.; Nes, I.F. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 1995, 47, 195–199. [Google Scholar] [PubMed]
- Ahmad, J.N.; Li, J.; Biedermannova, L.; Kuchar, M.; Sipova, H.; Semeradtova, A.; Cerny, J.; Petrokova, H.; Mikulecky, P.; Polinek, J.; et al. Novel high-affinity binders of human interferon gamma derived from albumin-binding domain of protein G. Proteins 2012, 80, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Mierau, I.; Kleerebezem, M. 10 years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2014, 47, 561-32. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Garcia, K.C. The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12. J. Mol. Biol. 2008, 382, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Eastman, P.; Pande, V.S. OpenMM: A Hardware Independent Framework for Molecular Simulations. Comput. Sci. Eng. 2015, 12, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.U.; Frick, I.M.; Nilsson, H.; Kraulis, P.J.; Hober, S.; Jonasson, P.; Linhult, M.; Nygren, P.A.; Uhlen, M.; Bjorck, L.; et al. Structure, specificity, and mode of interaction for bacterial albumin-binding modules. J. Biol. Chem. 2002, 277, 8114–8120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins 2013, 81, 2159–2166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kozakov, D.; Brenke, R.; Comeau, S.R.; Vajda, S. PIPER: An FFT-based protein docking program with pairwise potentials. Proteins 2006, 65, 392–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Strain, Plasmid, or Gene | Relevant Features or Sequence (5′–3′) | Reference or Source |
|---|---|---|
| Strain | ||
| E. coli | ||
| DH5α | endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR F- Φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK- mK+), λ– | Invitrogen |
| TOP10 | F– mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu) 7697 galU galK rpsL (StrR) endA1 nupG | Life technologies |
| BL21 λ(D3) | E. coli B F – dcm ompT hsdS (rB– mB–) gal λ(DE3) | [41] |
| L. lactis | ||
| NZ9000 | MG1363 nisRK ΔpepN | [42] |
| Plasmid | ||
| pNZ8148 | pSH71 derivative, PnisA, CmR, nisin-controlled expression | [42] |
| pSDLBA3b | pNZ8148 containing gene fusion of Usp45 signal peptide, B domain and cA | [32] |
| pET-ILP030 | pET28b containing a fusion gene of ILP030, tolA protein and AviTag consensus | [14] |
| pET-ILP317 | pET28b containing a fusion gene of ILP317, tolA protein and AviTag consensus | [14] |
| pET-ILP323 | pET28b containing a fusion gene of ILP323, tolA protein and AviTag consensus | [14] |
| pSD-ILP030 | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP030 and cA | This work |
| pSD-ILP317 | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP317 and cA | This work |
| pSD-ILP323 | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP323 and cA | This work |
| pSD-ADN23 | pNZ8148 containing gene fusion of Usp45 signal peptide, ADN23 and cA | [38] |
| pSD-ILP030-FLAG | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP030 and cA | This work |
| pSD-ILP317-FLAG | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP317 and cA | This work |
| pSD-ILP323-FLAG | pNZ8148 containing gene fusion of Usp45 signal peptide, ILP323 and cA | This work |
| pSD-ADN23-FLAG | pNZ8148 containing gene fusion of Usp45 signal peptide, ADN23 and cA | This work |
| pET-DH-TRX-p19 | pET28b containing a fusion gene of double-His-tag, Thioredoxin and p19 protein | This work |
| Primer | ||
| ILP030-F | TGGATCCTTAGCTGAAGCTAAAGTC | This work |
| ILP030-R | AGAATTCAGGTAAATTAGCTAAAATACG | This work |
| ILP317-R | AGAATTCAGGTAAAGGAGCTAAAATACTATC | This work |
| ILP323-R | AGAATTCAGGTAAACGAGCTAAAATAACATC | This work |
| Usp1-NcoI | ATAACCATGGCTAAAAAAAAGATTATCTCAGCTATTTTAATG | [32] |
| FLAG_Bam_R | GGATCCTTTATCATCGTCGTCTTTATAATCAGCGTAAACACCTGACAACG | This work |
| 19-F-NheI | GGGCTAGCTAGCAGAGCTGTGCCTGGGGGC | This work |
| p19-R-XhoI | GCGCCTCGAGGGGACTCAGGGTTGCTGCTC | This work |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Škrlec, K.; Zadravec, P.; Hlavničková, M.; Kuchař, M.; Vaňková, L.; Petroková, H.; Křížová, L.; Černý, J.; Berlec, A.; Malý, P. p19-Targeting ILP Protein Blockers of IL-23/Th-17 Pro-Inflammatory Axis Displayed on Engineered Bacteria of Food Origin. Int. J. Mol. Sci. 2018, 19, 1933. https://doi.org/10.3390/ijms19071933
Škrlec K, Zadravec P, Hlavničková M, Kuchař M, Vaňková L, Petroková H, Křížová L, Černý J, Berlec A, Malý P. p19-Targeting ILP Protein Blockers of IL-23/Th-17 Pro-Inflammatory Axis Displayed on Engineered Bacteria of Food Origin. International Journal of Molecular Sciences. 2018; 19(7):1933. https://doi.org/10.3390/ijms19071933
Chicago/Turabian StyleŠkrlec, Katja, Petra Zadravec, Marie Hlavničková, Milan Kuchař, Lucie Vaňková, Hana Petroková, Lucie Křížová, Jiří Černý, Aleš Berlec, and Petr Malý. 2018. "p19-Targeting ILP Protein Blockers of IL-23/Th-17 Pro-Inflammatory Axis Displayed on Engineered Bacteria of Food Origin" International Journal of Molecular Sciences 19, no. 7: 1933. https://doi.org/10.3390/ijms19071933