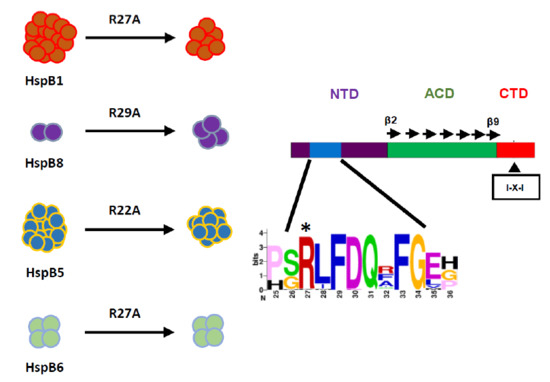
The Role of the Arginine in the Conserved N-Terminal Domain RLFDQxFG Motif of Human Small Heat Shock Proteins HspB1, HspB4, HspB5, HspB6, and HspB8
Abstract
:1. Introduction
2. Results
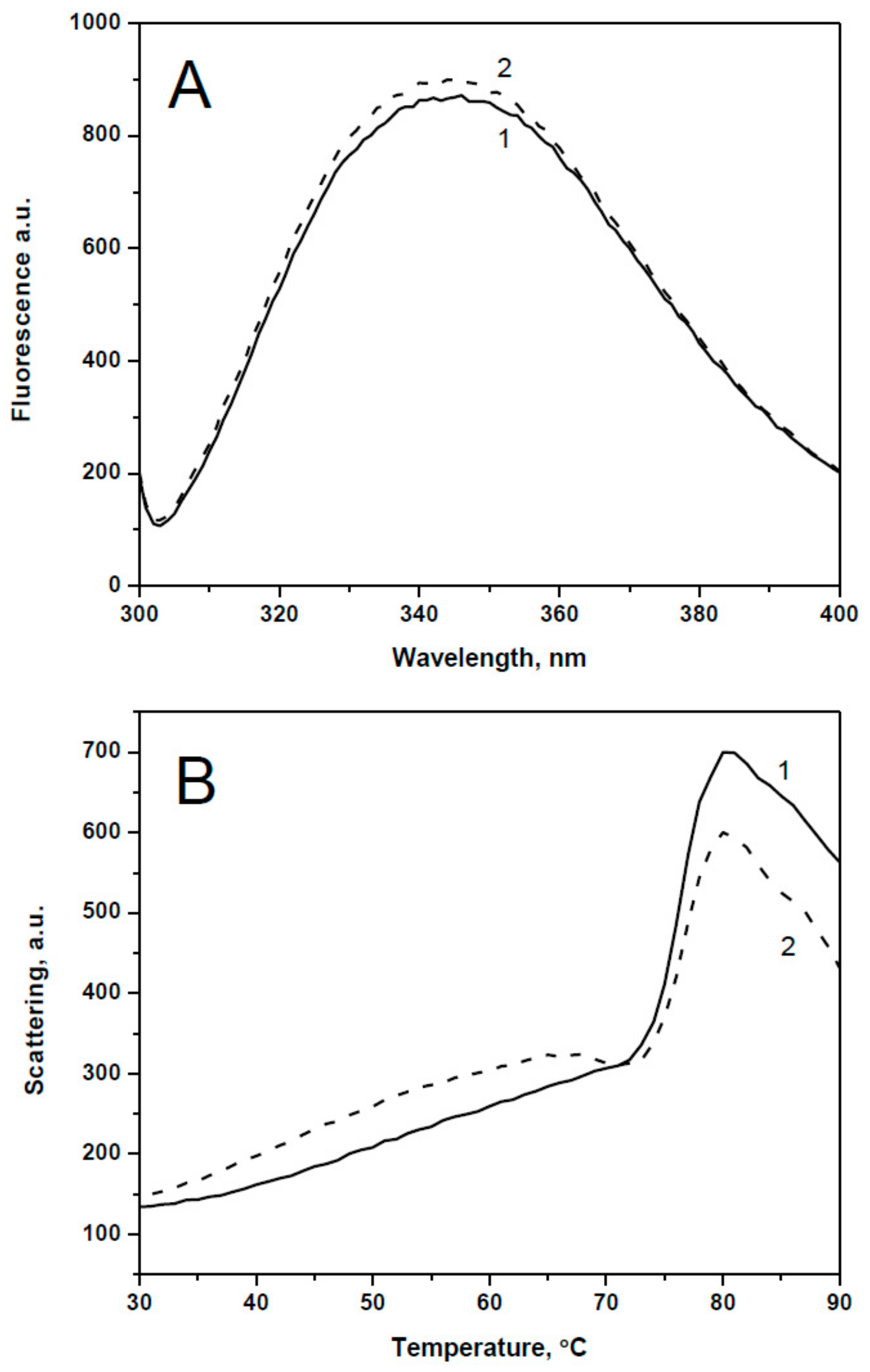
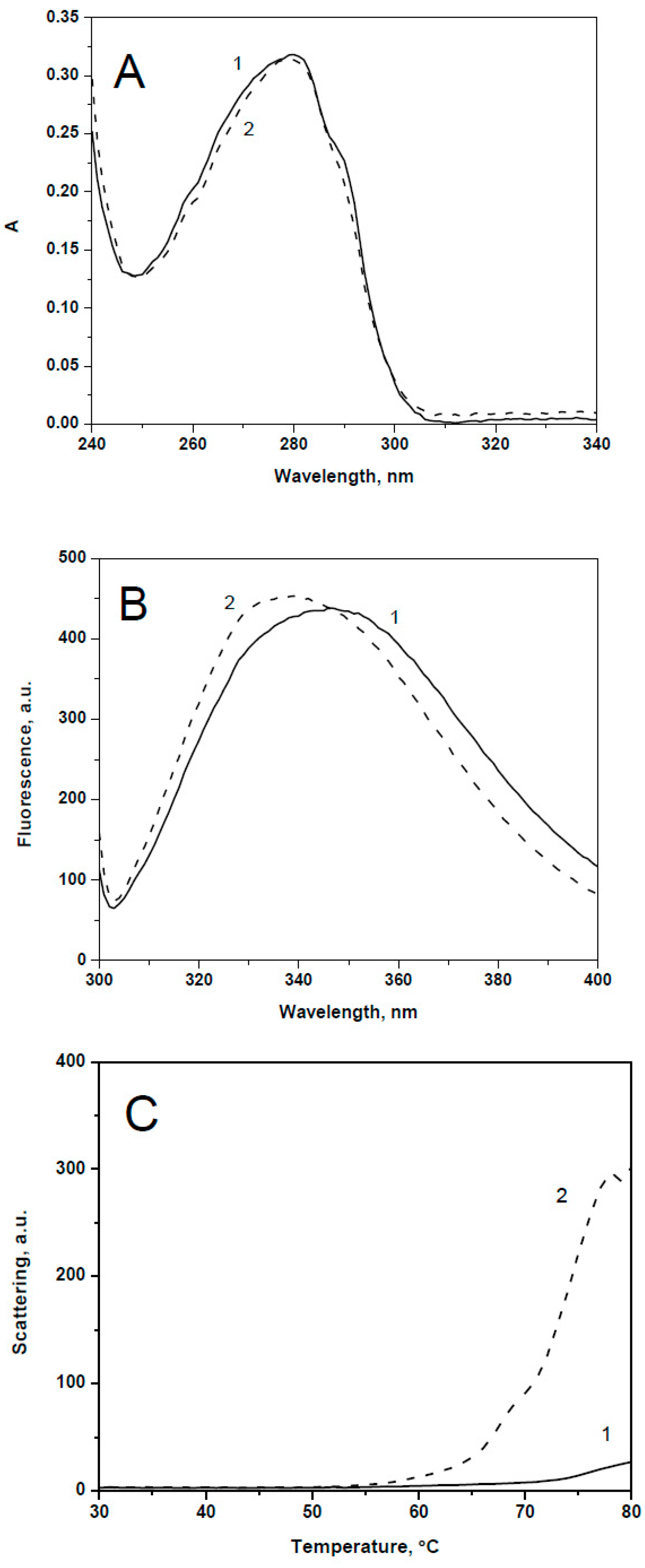
2.1. Spectral Properties and Thermal Stability of sHsps and Their Point Mutants
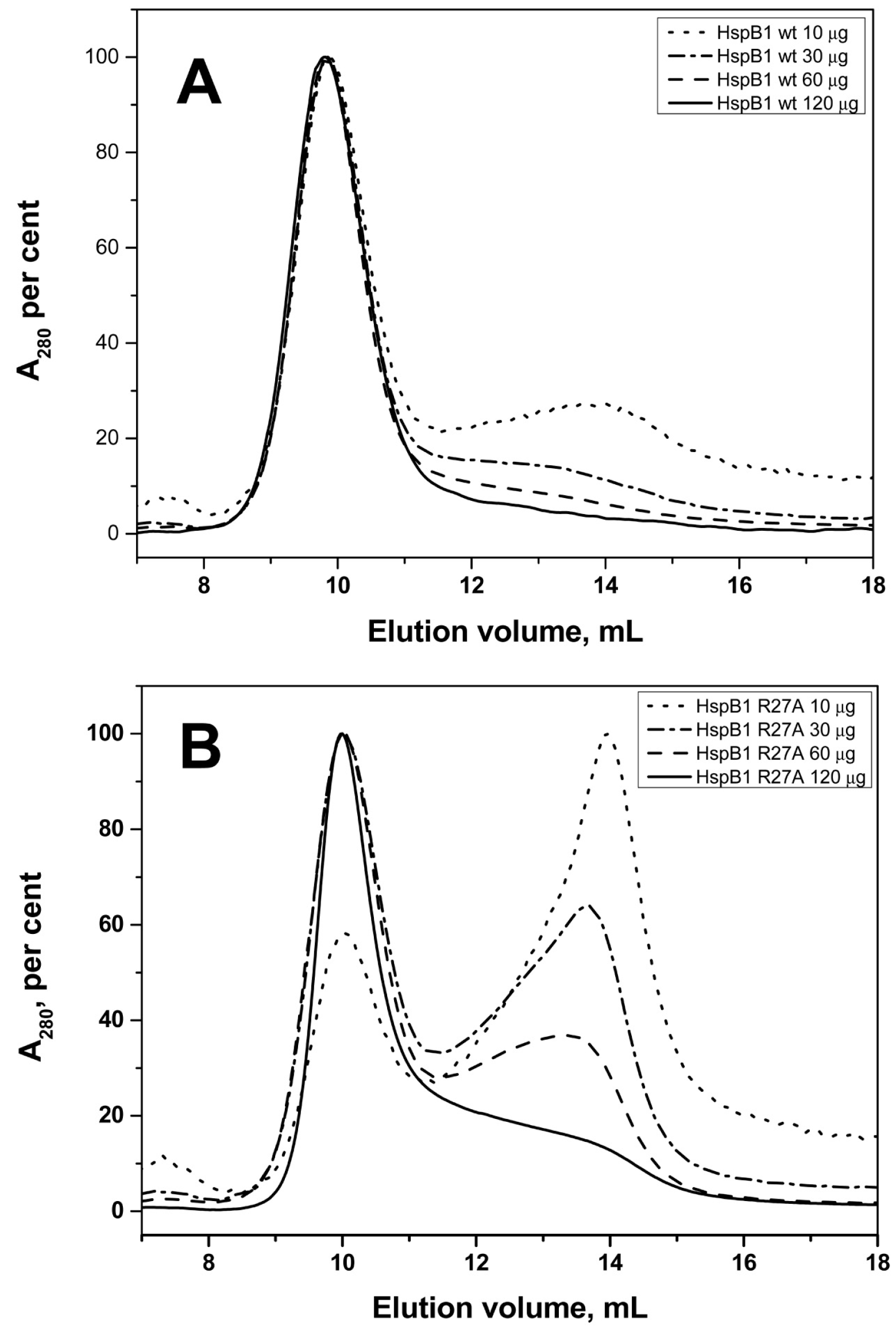
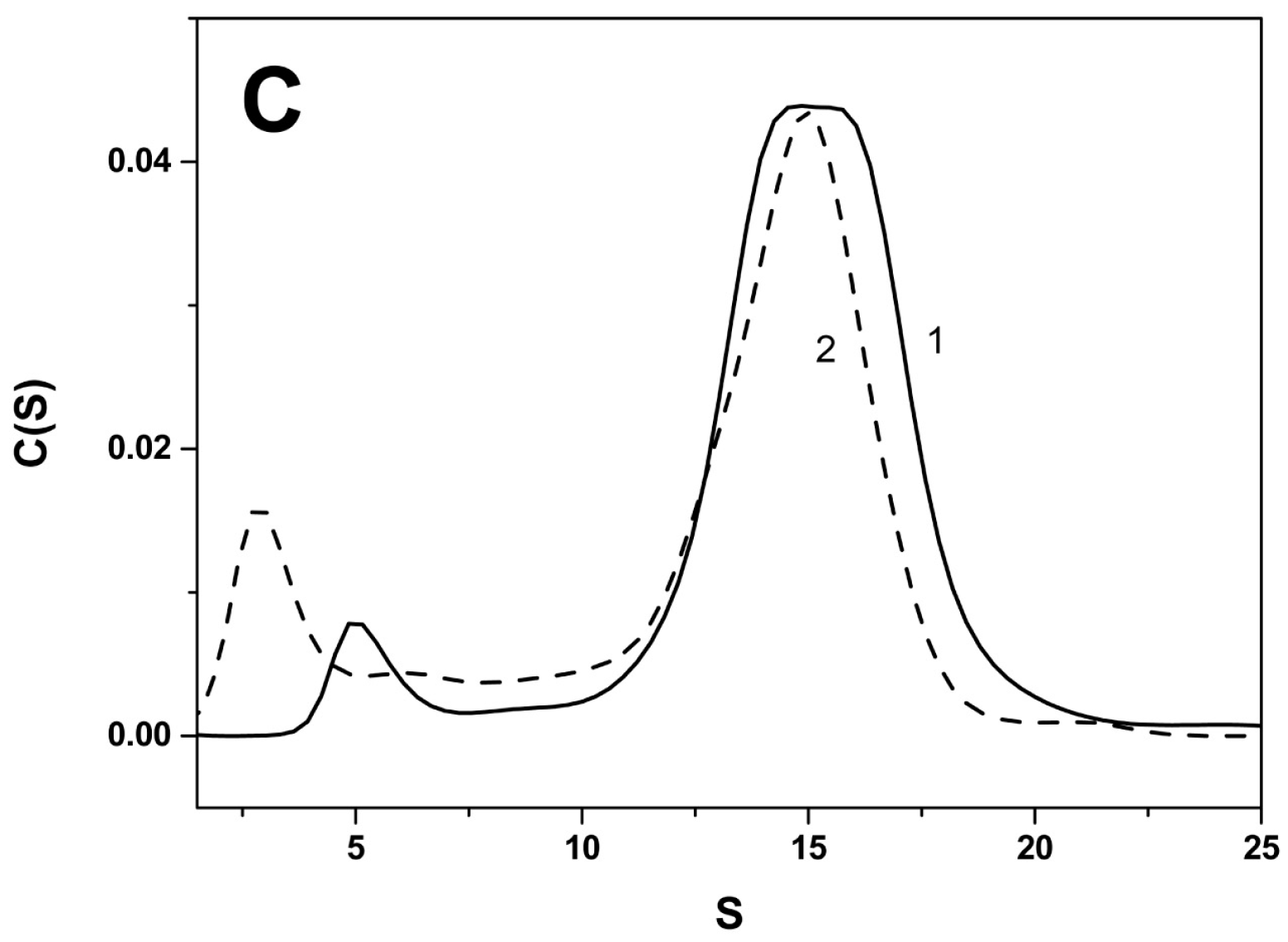
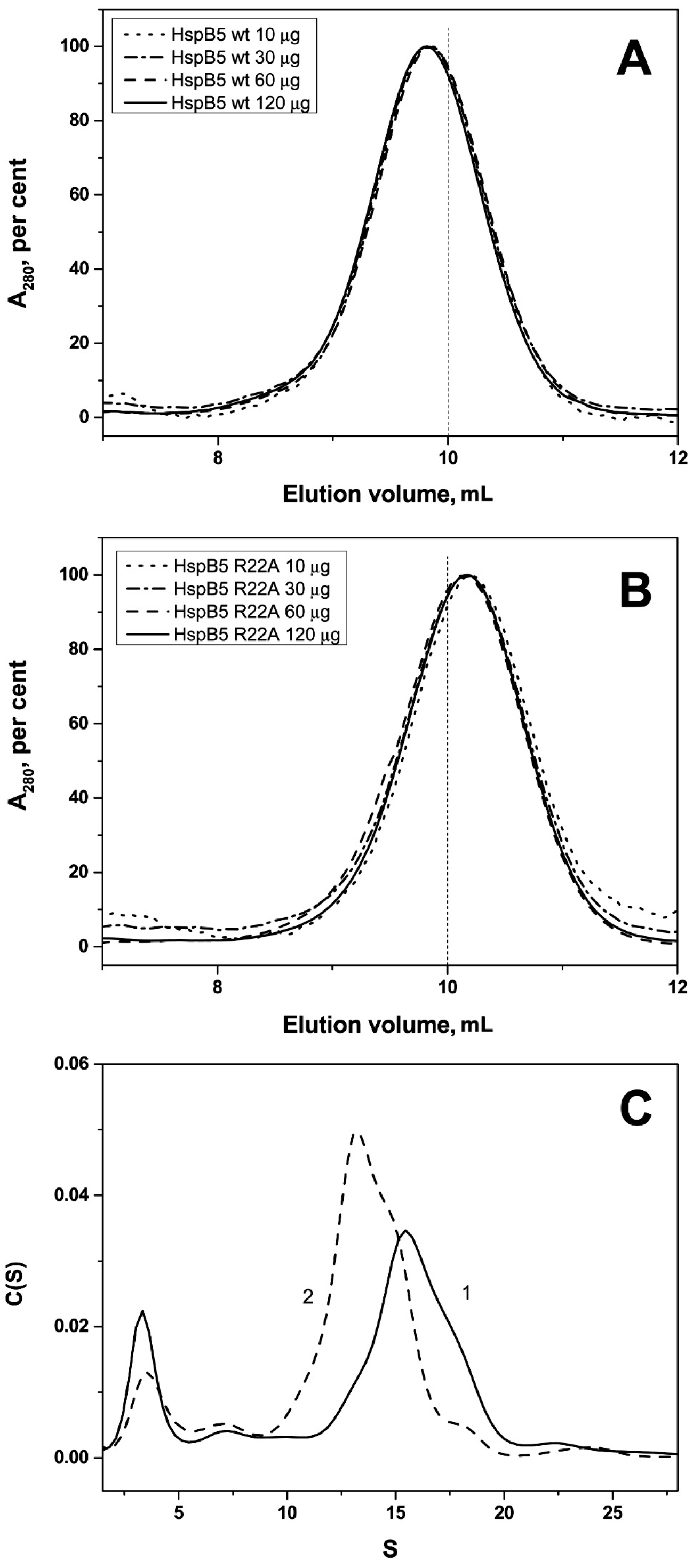
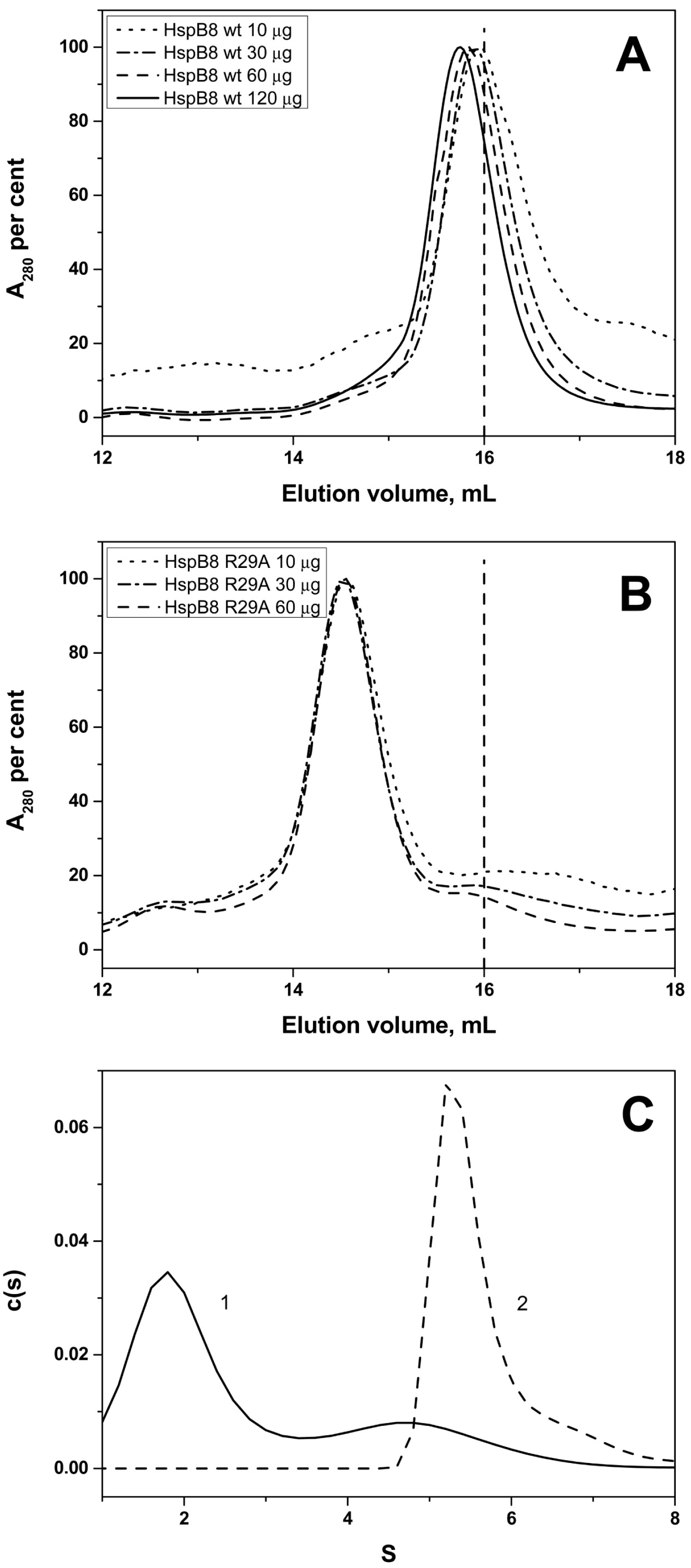
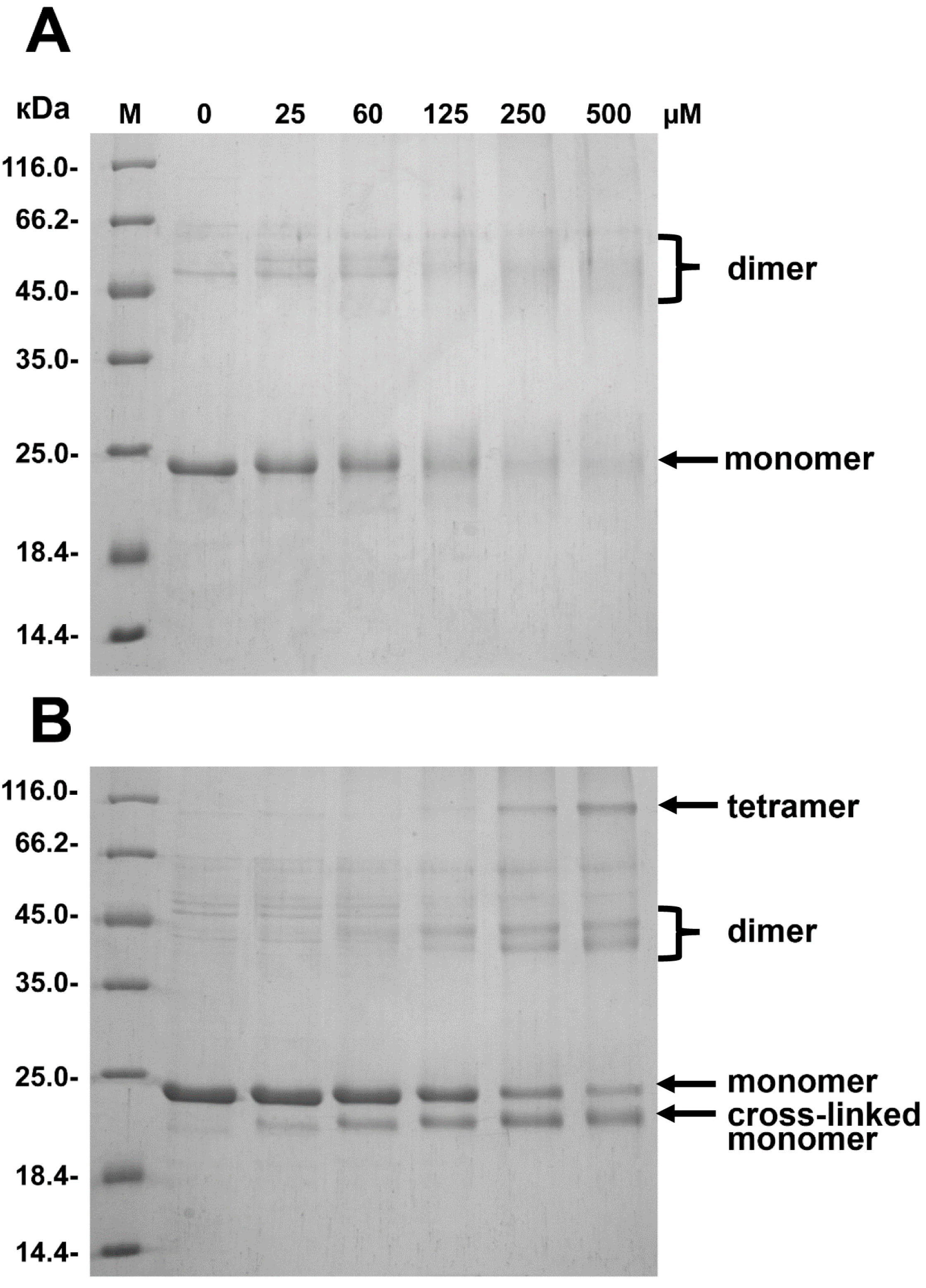
2.2. Effect of Arg/Ala Replacement on sHsp Quaternary Structure
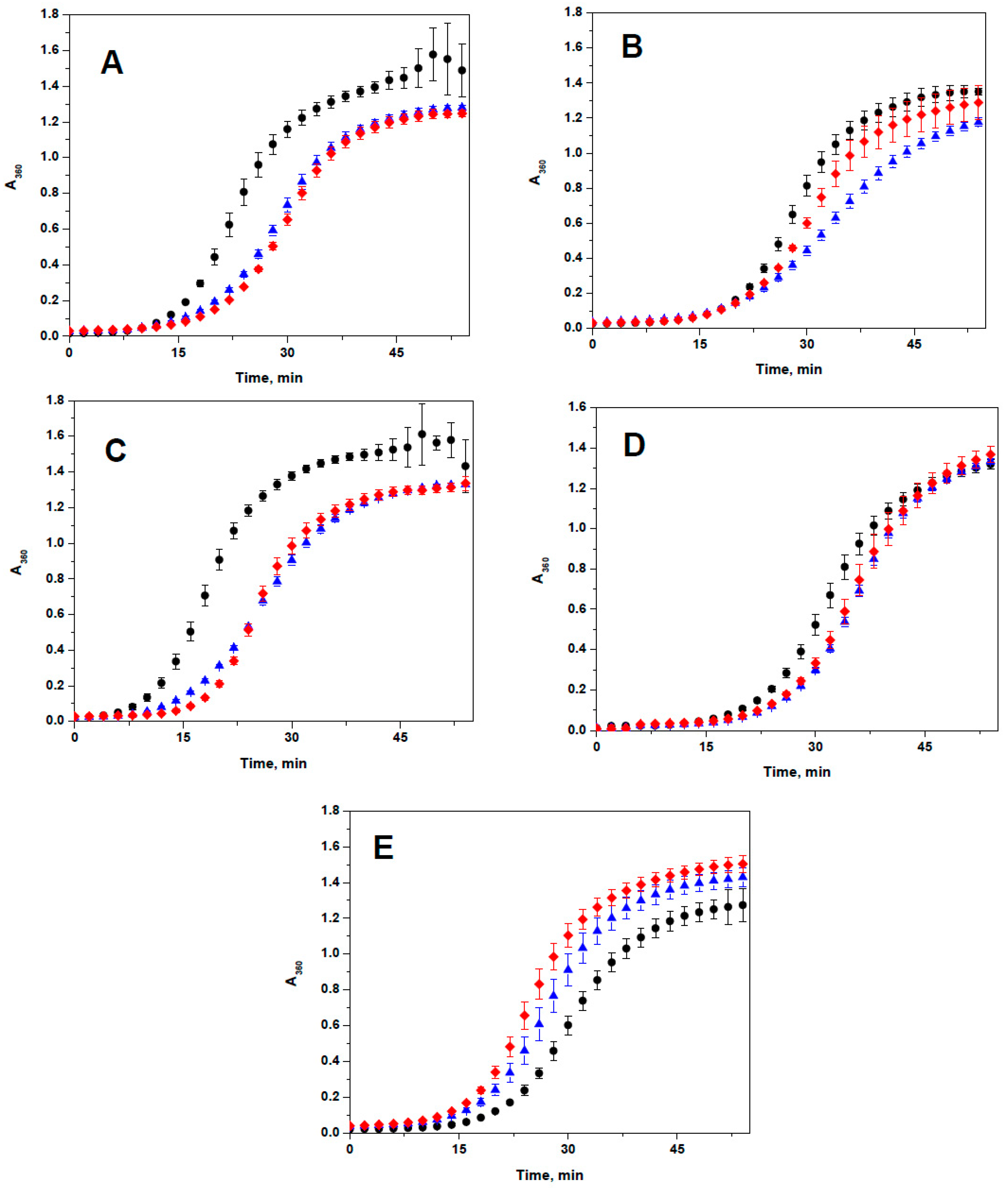
2.3. Lack of the Effect of Arg/Ala Replacement on Chaperone-Like Activity
3. Discussion
4. Materials and Methods
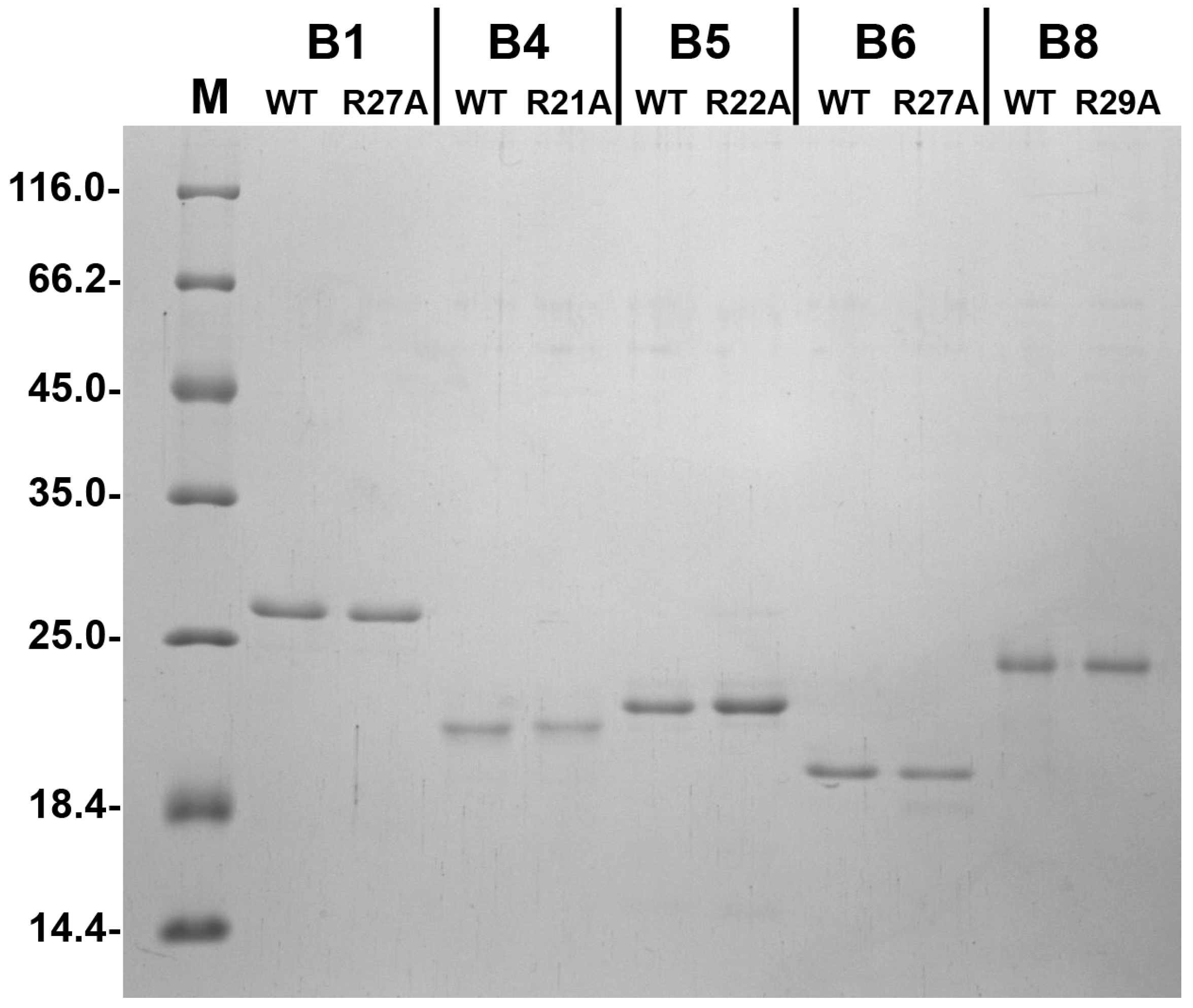
4.1. Proteins
4.2. Fluorescence Spectroscopy and Light Scattering
4.3. Size-Exclusion Chromatography
4.4. Analytical Ultracentrifugation
4.5. SEC-SAXS Studies
4.6. Crosslinking of HspB8
4.7. Chaperone-Like Activity
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kriehuber, T.; Rattei, T.; Weinmaier, T.; Bepperling, A.; Haslbeck, M.; Buchner, J. Independent evolution of the core domain and its flanking sequences in small heat shock proteins. FASEB J. 2010, 24, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Maaroufi, H.; Tanguay, R.M. Analysis and phylogeny of small heat shock proteins from marine viruses and their cyanobacteria host. PLoS ONE 2013, 8, e81207. [Google Scholar] [CrossRef] [PubMed]
- Slingsby, C.; Wistow, G.J. Functions of crystallins in and out of lens: Roles in elongated and post-mitotic cells. Prog. Biophys. Mol. Biol. 2014, 115, 52–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 2015, 1854, 291–319. [Google Scholar] [CrossRef] [PubMed]
- Treweek, T.M.; Meehan, S.; Ecroyd, H.; Carver, J.A. Small heat-shock proteins: Important players in regulating cellular proteostasis. Cell. Mol. Life Sci. 2015, 72, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Poulain, P.; Gelly, J.C.; Flatters, D. Detection and architecture of small heat shock protein monomers. PLoS ONE 2010, 5, e9990. [Google Scholar] [CrossRef] [PubMed]
- Hochberg, G.K.; Benesch, J.L. Dynamical structure of αb-crystallin. Prog. Biophys. Mol. Biol. 2014, 115, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Delbecq, S.P.; Rosenbaum, J.C.; Klevit, R.E. A mechanism of subunit recruitment in human small heat shock protein oligomers. Biochemistry 2015, 54, 4276–4284. [Google Scholar] [CrossRef] [PubMed]
- Haslbeck, M.; Peschek, J.; Buchner, J.; Weinkauf, S. Structure and function of α-crystallins: Traversing from in vitro to in vivo. Biochim. Biophys. Acta 2016, 1860, 149–166. [Google Scholar] [CrossRef] [PubMed]
- Sobott, F.; Benesch, J.L.; Vierling, E.; Robinson, C.V. Subunit exchange of multimeric protein complexes. Real-time monitoring of subunit exchange between small heat shock proteins by using electrospray mass spectrometry. J. Biol. Chem. 2002, 277, 38921–38929. [Google Scholar] [CrossRef] [PubMed]
- Benesch, J.L.; Ayoub, M.; Robinson, C.V.; Aquilina, J.A. Small heat shock protein activity is regulated by variable oligomeric substructure. J. Biol. Chem. 2008, 283, 28513–28517. [Google Scholar] [CrossRef] [PubMed]
- Jovcevski, B.; Kelly, M.A.; Rote, A.P.; Berg, T.; Gastall, H.Y.; Benesch, J.L.; Aquilina, J.A.; Ecroyd, H. Phosphomimics destabilize hsp27 oligomeric assemblies and enhance chaperone activity. Chem. Biol. 2015, 22, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Benndorf, R.; Hayess, K.; Ryazantsev, S.; Wieske, M.; Behlke, J.; Lutsch, G. Phosphorylation and supramolecular organization of murine small heat shock protein hsp25 abolish its actin polymerization-inhibiting activity. J. Biol. Chem. 1994, 269, 20780–20784. [Google Scholar] [PubMed]
- Aquilina, J.A.; Watt, S.J. The N-terminal domain of αb-crystallin is protected from proteolysis by bound substrate. Biochem. Biophys. Res. Commun. 2007, 353, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. Large potentials of small heat shock proteins. Physiol. Rev. 2011, 91, 1123–1159. [Google Scholar] [CrossRef] [PubMed]
- Peschek, J.; Braun, N.; Rohrberg, J.; Back, K.C.; Kriehuber, T.; Kastenmuller, A.; Weinkauf, S.; Buchner, J. Regulated structural transitions unleash the chaperone activity of αb-crystallin. Proc. Natl. Acad. Sci. USA 2013, 110, E3780–E3789. [Google Scholar] [CrossRef] [PubMed]
- Dephoure, N.; Zhou, C.; Villen, J.; Beausoleil, S.A.; Bakalarski, C.E.; Elledge, S.J.; Gygi, S.P. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 2008, 105, 10762–10767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heirbaut, M.; Strelkov, S.V.; Weeks, S.D. Everything but the ACD, Functional Conservation of the Non-conserved Terminal Regions in sHSPs. In The Big Book on Small Heat Shock Proteins; Tanguay, R.M., Hightower, L.E., Eds.; Springer International Publishing: Basel, Switzerland, 2015; pp. 197–227. [Google Scholar]
- Sudnitsyna, M.V.; Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. The role of intrinsically disordered regions in the structure and functioning of small heat shock proteins. Curr. Protein Pept. Sci. 2012, 13, 76–85. [Google Scholar] [CrossRef] [PubMed]
- Hanazono, Y.; Takeda, K.; Oka, T.; Abe, T.; Tomonari, T.; Akiyama, N.; Aikawa, Y.; Yohda, M.; Miki, K. Nonequivalence observed for the 16-meric structure of a small heat shock protein, sphsp16.0, from schizosaccharomyces pombe. Structure 2013, 21, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Van Montfort, R.L.; Basha, E.; Friedrich, K.L.; Slingsby, C.; Vierling, E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat. Struct. Biol. 2001, 8, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Merck, K.B.; De Haard-Hoekman, W.A.; Oude Essink, B.B.; Bloemendal, H.; De Jong, W.W. Expression and aggregation of recombinant α a-crystallin and its two domains. Biochim. Biophys. Acta 1992, 1130, 267–276. [Google Scholar] [CrossRef]
- Bova, M.P.; McHaourab, H.S.; Han, Y.; Fung, B.K. Subunit exchange of small heat shock proteins. Analysis of oligomer formation of αa-crystallin and hsp27 by fluorescence resonance energy transfer and site-directed truncations. J. Biol. Chem. 2000, 275, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Heirbaut, M.; Beelen, S.; Strelkov, S.V.; Weeks, S.D. Dissecting the functional role of the N-terminal domain of the human small heat shock protein hspb6. PLoS ONE 2014, 9, e105892. [Google Scholar] [CrossRef] [PubMed]
- Pasta, S.Y.; Raman, B.; Ramakrishna, T.; Rao Ch, M. Role of the conserved srlfdqffg region of α-crystallin, a small heat shock protein. Effect on oligomeric size, subunit exchange, and chaperone-like activity. J. Biol. Chem. 2003, 278, 51159–51166. [Google Scholar] [CrossRef] [PubMed]
- Kumar, L.V.; Rao, C.M. Domain swapping in human α a and α b crystallins affects oligomerization and enhances chaperone-like activity. J. Biol. Chem. 2000, 275, 22009–22013. [Google Scholar] [CrossRef] [PubMed]
- Heirbaut, M.; Lermyte, F.; Martin, E.M.; Beelen, S.; Sobott, F.; Strelkov, S.V.; Weeks, S.D. Specific sequences in the n-terminal domain of human small heat-shock protein hspb6 dictate preferential hetero-oligomerization with the orthologue hspb1. J. Biol. Chem. 2017, 292, 9944–9957. [Google Scholar] [CrossRef] [PubMed]
- Santhoshkumar, P.; Sharma, K.K. Phe71 is essential for chaperone-like function in α a-crystallin. J. Biol. Chem. 2001, 276, 47094–47099. [Google Scholar] [CrossRef] [PubMed]
- Muranova, L.K.; Weeks, S.D.; Strelkov, S.V.; Gusev, N.B. Characterization of mutants of human small heat shock protein hspb1 carrying replacements in the n-terminal domain and associated with hereditary motor neuron diseases. PLoS ONE 2015, 10, e0126248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irobi, J.; Van Impe, K.; Seeman, P.; Jordanova, A.; Dierick, I.; Verpoorten, N.; Michalik, A.; De Vriendt, E.; Jacobs, A.; Van Gerwen, V.; et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat. Genet. 2004, 36, 597–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaoui, R.; Palmio, J.; Brewer, J.; Lek, M.; Needham, M.; Evila, A.; Hackman, P.; Jonson, P.H.; Penttila, S.; Vihola, A.; et al. Mutations in hspb8 causing a new phenotype of distal myopathy and motor neuropathy. Neurology 2016, 86, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Xiao, H.; Hu, Z.; Zhou, F.; Yang, B. Exploring the multifaceted roles of heat shock protein b8 (hspb8) in diseases. Eur. J. Cell Biol. 2018, 97, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.D.; Baranova, E.V.; Heirbaut, M.; Beelen, S.; Shkumatov, A.V.; Gusev, N.B.; Strelkov, S.V. Molecular structure and dynamics of the dimeric human small heat shock protein hspb6. J. Struct. Biol. 2014, 185, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Heirbaut, M.; Lermyte, F.; Martin, E.M.; Beelen, S.; Verschueren, T.; Sobott, F.; Strelkov, S.V.; Weeks, S.D. The preferential heterodimerization of human small heat shock proteins hspb1 and hspb6 is dictated by the n-terminal domain. Arch. Biochem. Biophys. 2016, 610, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.V.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Some properties of human small heat shock protein hsp22 (h11 or hspb8). Biochem. Biophys. Res. Commun. 2004, 315, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Rambo, R.P.; Tainer, J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 2013, 496, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delbecq, S.P.; Jehle, S.; Klevit, R. Binding determinants of the small heat shock protein, αb-crystallin: Recognition of the ixi motif. EMBO J. 2012, 31, 4587–4594. [Google Scholar] [CrossRef] [PubMed]
- Koteiche, H.A.; Chiu, S.; Majdoch, R.L.; Stewart, P.L.; McHaourab, H.S. Atomic models by cryo-em and site-directed spin labeling: Application to the n-terminal region of hsp16.5. Structure 2005, 13, 1165–1171. [Google Scholar] [CrossRef] [PubMed]
- Jehle, S.; Vollmar, B.S.; Bardiaux, B.; Dove, K.K.; Rajagopal, P.; Gonen, T.; Oschkinat, H.; Klevit, R.E. N-terminal domain of alphaB-crystallin provides a conformational switch for multimerization and structural heterogeneity. Proc. Natl. Acad. Sci. USA 2011, 108, 6409–6414. [Google Scholar] [CrossRef] [PubMed]
- McDonald, E.T.; Bortolus, M.; Koteiche, H.A.; McHaourab, H.S. Sequence, structure, and dynamic determinants of hsp27 (hspb1) equilibrium dissociation are encoded by the n-terminal domain. Biochemistry 2012, 51, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Rogalla, T.; Ehrnsperger, M.; Preville, X.; Kotlyarov, A.; Lutsch, G.; Ducasse, C.; Paul, C.; Wieske, M.; Arrigo, A.P.; Buchner, J.; et al. Regulation of hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor α by phosphorylation. J. Biol. Chem. 1999, 274, 18947–18956. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.; Yao, W.; Eiberg, H.; Kjaer, K.W.; Baggesen, K.; Hejtmancik, J.F.; Rosenberg, T. Genetic heterogeneity in microcornea-cataract: Five novel mutations in cryaa, crygd, and GJA8. Investig. Ophthalmol. Vis. Sci. 2007, 48, 3937–3944. [Google Scholar] [CrossRef] [PubMed]
- Devi, R.R.; Yao, W.; Vijayalakshmi, P.; Sergeev, Y.V.; Sundaresan, P.; Hejtmancik, J.F. Crystallin gene mutations in indian families with inherited pediatric cataract. Mol. Vis. 2008, 14, 1157–1170. [Google Scholar] [PubMed]
- Graw, J.; Klopp, N.; Illig, T.; Preising, M.N.; Lorenz, B. Congenital cataract and macular hypoplasia in humans associated with a de novo mutation in cryaa and compound heterozygous mutations in P. Graefes Arch. Clin. Exp. Ophthalmol. 2006, 244, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Laurie, K.J.; Dave, A.; Straga, T.; Souzeau, E.; Chataway, T.; Sykes, M.J.; Casey, T.; Teo, T.; Pater, J.; Craig, J.E.; et al. Identification of a novel oligomerization disrupting mutation in cryαa associated with congenital cataract in a south australian family. Hum. Mutat. 2013, 34, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Javadiyan, S.; Craig, J.E.; Souzeau, E.; Sharma, S.; Lower, K.M.; Pater, J.; Casey, T.; Hodson, T.; Burdon, K.P. Recurrent mutation in the crystallin α a gene associated with inherited paediatric cataract. BMC Res. Notes 2016, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.V.; Kasakov, A.S.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Structure and properties of k141e mutant of small heat shock protein hsp22 (hspb8, h11) that is expressed in human neuromuscular disorders. Arch. Biochem. Biophys. 2006, 454, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Sluchanko, N.N.; Chebotareva, N.A.; Gusev, N.B. Quaternary structure of human small heat shock protein hspb6 (hsp20) in crowded media modeled by trimethylamine n-oxide (tmao): Effect of protein phosphorylation. Biochimie 2015, 108, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Bukach, O.V.; Seit-Nebi, A.S.; Marston, S.B.; Gusev, N.B. Some properties of human small heat shock protein hsp20 (hspb6). Eur. J. Biochem. 2004, 271, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Chowdary, T.K.; Raman, B.; Ramakrishna, T.; Rao, C.M. Mammalian hsp22 is a heat-inducible small heat-shock protein with chaperone-like activity. Biochem. J. 2004, 381, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Villen, J.; Beausoleil, S.A.; Gerber, S.A.; Gygi, S.P. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA 2007, 104, 1488–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shemetov, A.A.; Seit-Nebi, A.S.; Gusev, N.B. Phosphorylation of human small heat shock protein hspb8 (hsp22) by erk1 protein kinase. Mol. Cell. Biochem. 2011, 355, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Mymrikov, E.V.; Daake, M.; Richter, B.; Haslbeck, M.; Buchner, J. The chaperone activity and substrate spectrum of human small heat shock proteins. J. Biol. Chem. 2017, 292, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Vos, M.J.; Kanon, B.; Kampinga, H.H. Hspb7 is a sc35 speckle resident small heat shock protein. Biochim. Biophys. Acta 2009, 1793, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Koteiche, H.A.; McHaourab, H.S.; Stewart, P.L. Cryoelectron microscopy and epr analysis of engineered symmetric and polydisperse hsp16.5 assemblies reveals determinants of polydispersity and substrate binding. J. Biol. Chem. 2006, 281, 40420–40428. [Google Scholar] [CrossRef] [PubMed]
- Van de Klundert, F.A.; Smulders, R.H.; Gijsen, M.L.; Lindner, R.A.; Jaenicke, R.; Carver, J.A.; de Jong, W.W. The mammalian small heat-shock protein hsp20 forms dimers and is a poor chaperone. Eur. J. Biochem. 1998, 258, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones 2012, 17, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lebowitz, J.; Lewis, M.S.; Schuck, P. Modern analytical ultracentrifugation in protein science: A tutorial review. Protein Sci. 2002, 11, 2067–2079. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Perez, J. Combined sampler robot and high-performance liquid chromatography: A fully automated system for biological small-angle x-ray scattering experiments at the synchrotron soleil swing beamline. J. Appl. Crystallogr. 2009, 42, 892–900. [Google Scholar] [CrossRef]
- Alshammari, E.M.; Khan, S.; Jawed, A.; Adnan, M.; Khan, M.; Nabi, G.; Lohani, M.; Haque, S. Optimization of extraction parameters for enhanced production of ovotransferrin from egg white for antimicrobial applications. BioMed Res. Int. 2015, 2015, 934512. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.W.; Holbrook, J.J. Differences in the protein fluorecence of the two iron(iii)-binding sites of ovotransferrin. Biochem. J. 1975, 145, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.D.; Muranova, L.K.; Heirbaut, M.; Beelen, S.; Strelkov, S.V.; Gusev, N.B. Characterization of human small heat shock protein hspb1 α-crystallin domain localized mutants associated with hereditary motor neuron diseases. Sci. Rep. 2018, 8, 688. [Google Scholar] [CrossRef] [PubMed]

| Protein | Monomeric Mass, kDa | Apparent Mass/No. of Monomers WT | Apparent Mass/No. of Monomers Mutant |
|---|---|---|---|
| HspB1 | 22.8 | 540/~24 | 500/~22 + 80/~4 * |
| HspB4 | 19.9 | 590/~30 | 590/~30 |
| HspB5 | 20.2 | 540/~27 | 460/~23 |
| HspB6 | 17.1 | 50/~3 ** | 50/~3 |
| HspB8 | 21.6 | 33/~2 *** | 60/~3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shatov, V.M.; Weeks, S.D.; Strelkov, S.V.; Gusev, N.B. The Role of the Arginine in the Conserved N-Terminal Domain RLFDQxFG Motif of Human Small Heat Shock Proteins HspB1, HspB4, HspB5, HspB6, and HspB8. Int. J. Mol. Sci. 2018, 19, 2112. https://doi.org/10.3390/ijms19072112
Shatov VM, Weeks SD, Strelkov SV, Gusev NB. The Role of the Arginine in the Conserved N-Terminal Domain RLFDQxFG Motif of Human Small Heat Shock Proteins HspB1, HspB4, HspB5, HspB6, and HspB8. International Journal of Molecular Sciences. 2018; 19(7):2112. https://doi.org/10.3390/ijms19072112
Chicago/Turabian StyleShatov, Vladislav M., Stephen D. Weeks, Sergei V. Strelkov, and Nikolai B. Gusev. 2018. "The Role of the Arginine in the Conserved N-Terminal Domain RLFDQxFG Motif of Human Small Heat Shock Proteins HspB1, HspB4, HspB5, HspB6, and HspB8" International Journal of Molecular Sciences 19, no. 7: 2112. https://doi.org/10.3390/ijms19072112