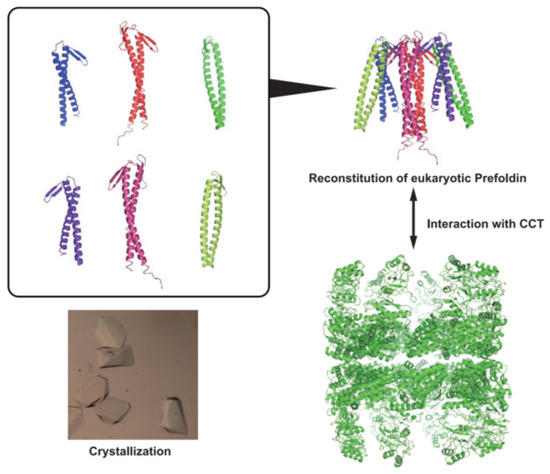
Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum
Abstract
:1. Introduction
2. Results
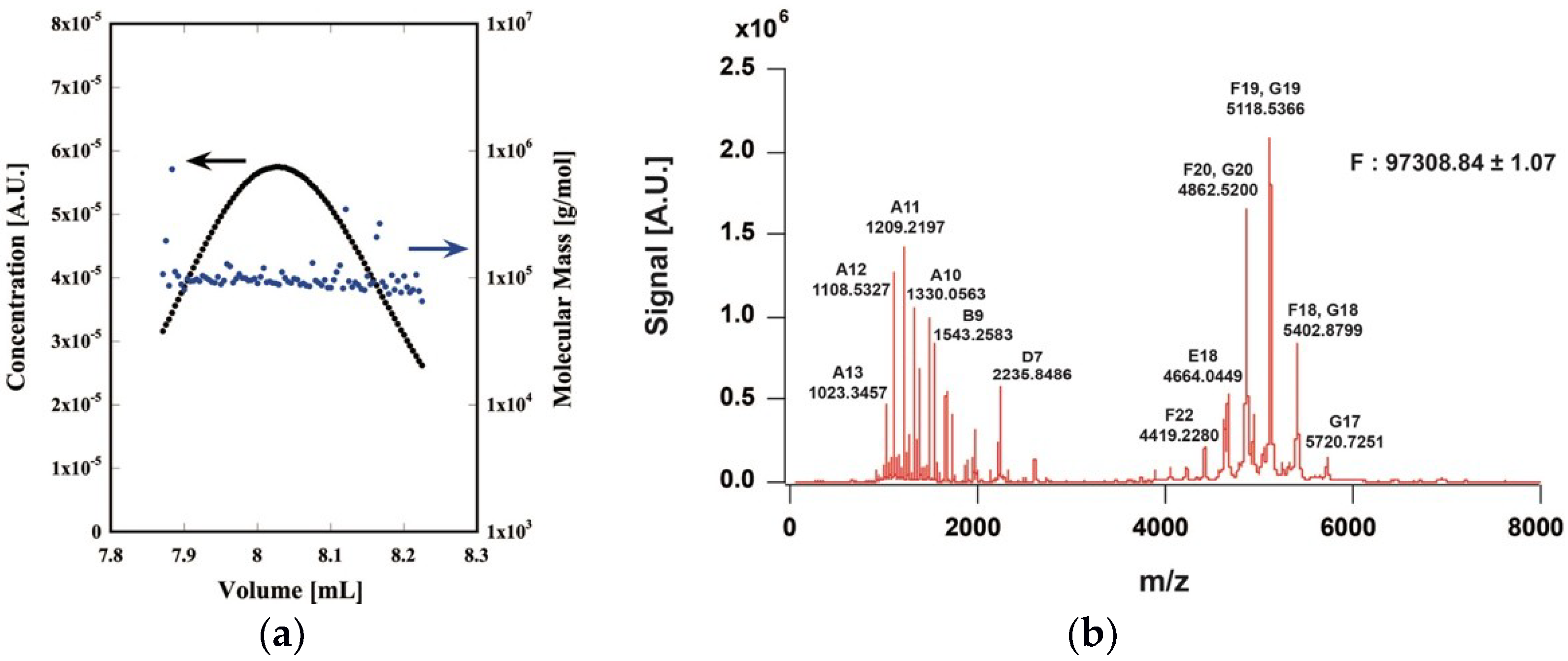
2.1. Expression and Purification of the CtPFD
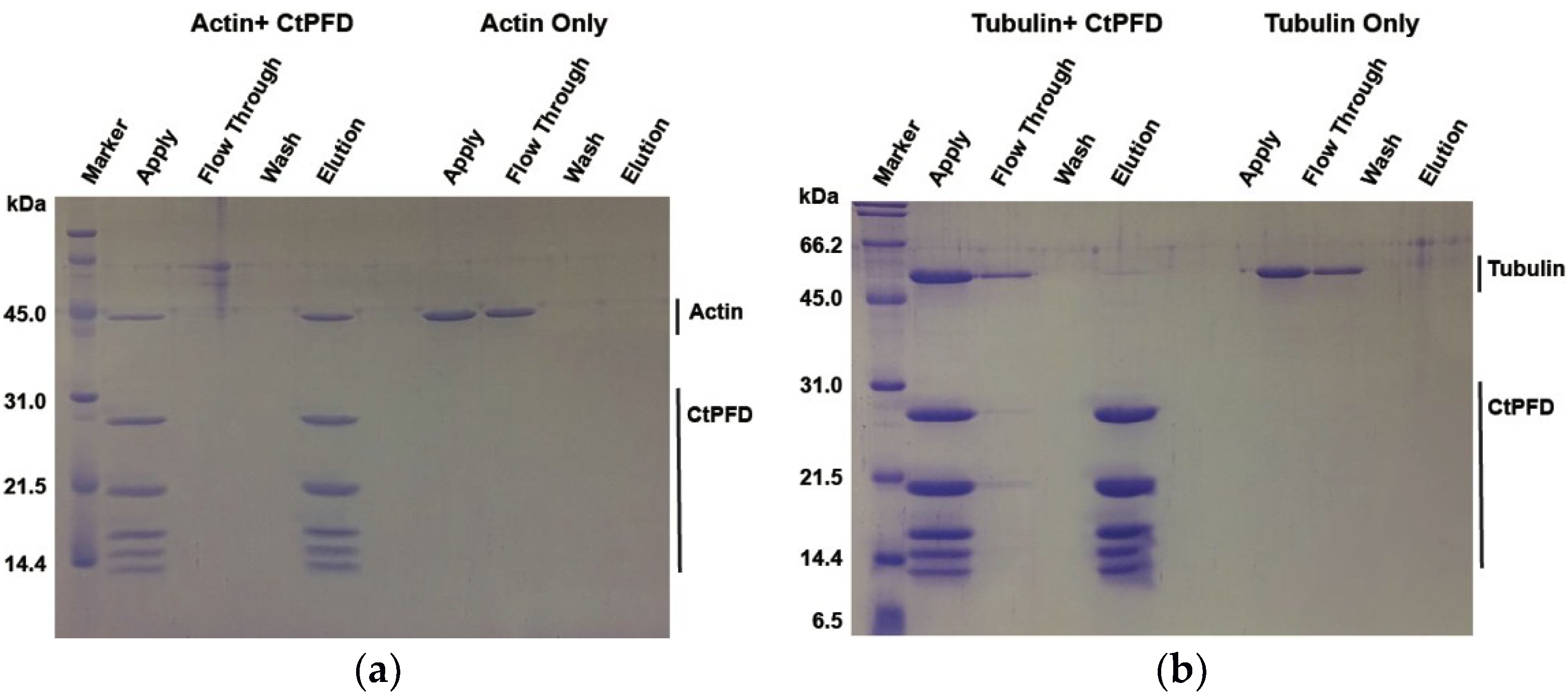
2.2. Interaction with Substrates
2.3. Interaction with CtCCT
2.4. Actin Transfer to CtCCT
2.5. Crystallization
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Fungi, Proteins, and Reagents
4.2. Cloning, Expression, and Purification of CtPFD
4.3. Size Exclusion Chromatography-Multiangle Light Scattering (SEC-MALS)
4.4. Native Mass Spectrometry
4.5. Protection of Thermal Aggregation of CS
4.6. Pull-Down Assay
4.7. Analysis of CtPFD–CtCCT Interaction by SPR
4.8. HS-AFM Observation
4.9. Transmission Electron Microscopy
4.10. Actin Delivery Assay
4.11. Crystallization and X-ray Data Collection
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCT | Chaperonin containing TCP-1 |
| PFD | Prefoldin |
| TRiC | TCP-1 ring complex |
| CtCCD | CCT from Chaetomium thermophilum |
| CtPFD | PFD from Chaetomium thermophilum |
| PFD1 | Subunit 1 of CtPFD |
| PFD2 | Subunit 2 of CtPFD |
| PFD3 | Subunit 3 of CtPFD |
| PFD4 | Subunit 4 of CtPFD |
| PFD5 | Subunit 5 of CtPFD |
| PFD6 | Subunit 6 of CtPFD |
| CS | Porcine heart citrate synthase |
| SPR | Surface plasmon resonance |
| HS-AFM | High-speed atomic force microscopy |
References
- Hartl, F.U.; Hayer-Hartl, M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 2002, 295, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, I.; Essen, L.O.; Baumeister, W. Group II chaperonins: New TRiC(k)s and turns of a protein folding machine. J. Mol. Biol. 1999, 293, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Horwich, A.L.; Fenton, W.A.; Chapman, E.; Farr, G.W. Two families of chaperonin: Physiology and mechanism. Annu. Rev. Cell. Dev. Biol. 2007, 23, 115–145. [Google Scholar] [CrossRef] [PubMed]
- Geissler, S.; Siegers, K.; Schiebel, E. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 1998, 17, 952–966. [Google Scholar] [CrossRef] [PubMed]
- Vainberg, I.E.; Lewis, S.A.; Rommelaere, H.; Ampe, C.; Vandekerckhove, J.; Klein, H.L.; Cowan, N.J. Prefoldin, a chaperone that delivers unfolded proteins to cytosolic chaperonin. Cell 1998, 93, 863–873. [Google Scholar] [CrossRef]
- Leroux, M.R.; Fandrich, M.; Klunker, D.; Siegers, K.; Lupas, A.N.; Brown, J.R.; Schiebel, E.; Dobson, C.M.; Hartl, F.U. MtGimC, a novel archaeal chaperone related to the eukaryotic chaperonin cofactor GimC/prefoldin. EMBO J. 1999, 18, 6730–6743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okochi, M.; Yoshida, T.; Maruyama, T.; Kawarabayasi, Y.; Kikuchi, H.; Yohda, M. Pyrococcus prefoldin stabilizes protein-folding intermediates and transfers them to chaperonins for correct folding. Biochem. Biophys. Res. Commun. 2002, 291, 769–774. [Google Scholar] [CrossRef] [PubMed]
- Okochi, M.; Nomura, T.; Zako, T.; Arakawa, T.; Iizuka, R.; Ueda, H.; Funatsu, T.; Leroux, M.; Yohda, M. Kinetics and binding sites for interaction of the prefoldin with a group II chaperonin: Contiguous non-native substrate and chaperonin binding sites in the archaeal prefoldin. J. Biol. Chem. 2004, 279, 31788–31795. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, A.; Kida, H.; Miyata, Y.; Ide, N.; Yonezawa, A.; Arakawa, T.; Iizuka, R.; Noguchi, K.; Kita, A.; Odaka, M.; et al. Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins. J. Mol. Biol. 2008, 376, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Siegert, R.; Leroux, M.R.; Scheufler, C.; Hartl, F.U.; Moarefi, I. Structure of the molecular chaperone prefoldin: Unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell 2000, 103, 621–632. [Google Scholar] [CrossRef]
- Zako, T.; Iizuka, R.; Okochi, M.; Nomura, T.; Ueno, T.; Tadakuma, H.; Yohda, M.; Funatsu, T. Facilitated release of substrate protein from prefoldin by chaperonin. FEBS Lett. 2005, 579, 3718–3724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zako, T.; Murase, Y.; Iizuka, R.; Yoshida, T.; Kanzaki, T.; Ide, N.; Maeda, M.; Funatsu, T.; Yohda, M. Localization of prefoldin interaction sites in the hyperthermophilic group II chaperonin and correlations between binding rate and protein transfer rate. J. Mol. Biol. 2006, 364, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Zako, T.; Sahlan, M.; Fujii, S.; Yamamoto, Y.Y.; Tai, P.T.; Sakai, K.; Maeda, M.; Yohda, M. Contribution of the C-Terminal Region of a Group II Chaperonin to its Interaction with Prefoldin and Substrate Transfer. J. Mol. Biol. 2016, 428, 2405–2417. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Takahashi-Niki, K.; Takekoshi, Y.; Shimizu, T.; Kitaura, H.; Maita, H.; Iguchi-Ariga, S.M.; Ariga, H. Prefoldin plays a role as a clearance factor in preventing proteasome inhibitor-induced protein aggregation. J. Biol. Chem. 2013, 288, 27764–27776. [Google Scholar] [CrossRef] [PubMed]
- Simons, C.T.; Staes, A.; Rommelaere, H.; Ampe, C.; Lewis, S.A.; Cowan, N.J. Selective contribution of eukaryotic prefoldin subunits to actin and tubulin binding. J. Biol. Chem. 2004, 279, 4196–4203. [Google Scholar] [CrossRef] [PubMed]
- Martin-Benito, J.; Boskovic, J.; Gomez-Puertas, P.; Carrascosa, J.L.; Simons, C.T.; Lewis, S.A.; Bartolini, F.; Cowan, N.J.; Valpuesta, J.M. Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 2002, 21, 6377–6386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aikawa, Y.; Kida, H.; Nishitani, Y.; Miki, K. Expression, purification, crystallization and X-ray diffraction studies of the molecular chaperone prefoldin from Homo sapiens. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 1189–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez de Diego, R.; Ortiz-Lombardia, M.; Bravo, J. Crystallization and preliminary X-ray diffraction analysis of the beta subunit Yke2 of the Gim complex from Saccharomyces cerevisiae. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2008, 64, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.Y.; Uno, Y.; Sha, E.; Ikegami, K.; Ishii, N.; Dohmae, N.; Sekiguchi, H.; Sasaki, Y.C.; Yohda, M. Asymmetry in the function and dynamics of the cytosolic group II chaperonin CCT/TRiC. PLoS ONE 2017, 12, e0176054. [Google Scholar] [CrossRef] [PubMed]
- Amlacher, S.; Sarges, P.; Flemming, D.; van Noort, V.; Kunze, R.; Devos, D.P.; Arumugam, M.; Bork, P.; Hurt, E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011, 146, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Bock, T.; Chen, W.H.; Ori, A.; Malik, N.; Silva-Martin, N.; Huerta-Cepas, J.; Powell, S.T.; Kastritis, P.L.; Smyshlyaev, G.; Vonkova, I.; et al. An integrated approach for genome annotation of the eukaryotic thermophile Chaetomium thermophilum. Nucleic Acids Res. 2014, 42, 13525–13533. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Mimori, K.; Nishida, N.; Tokuoka, M.; Akita, H.; Sekimoto, M.; Doki, Y.; Mori, M. Abnormal expression of PFDN4 in colorectal cancer: a novel marker for prognosis. Ann. Surg. Oncol. 2010, 17, 3030–3036. [Google Scholar] [CrossRef] [PubMed]
- Lopez, V.; Gonzalez-Peramato, P.; Suela, J.; Serrano, A.; Algaba, F.; Cigudosa, J.C.; Vidal, A.; Bellmunt, J.; Heredero, O.; Sanchez-Carbayo, M. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J. Transl. Med. 2013, 11, 182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Zhao, J.; Yang, X.; Guan, S.; Feng, H.; Han, D.; Lu, J.; Ou, B.; Jin, R.; Sun, J.; et al. PFDN1, an indicator for colorectal cancer prognosis, enhances tumor cell proliferation and motility through cytoskeletal reorganization. Med. Oncol. 2015, 32, 264. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Shi, W.; Tang, Y.; Liu, Y.; He, K.; Hu, Y.; Li, J.; Yang, Y.; Song, J. Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene 2017, 36, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Millan-Zambrano, G.; Chavez, S. Nuclear functions of prefoldin. Open Biol. 2014, 4, 140085. [Google Scholar] [CrossRef] [PubMed]
- Gstaiger, M.; Luke, B.; Hess, D.; Oakeley, E.J.; Wirbelauer, C.; Blondel, M.; Vigneron, M.; Peter, M.; Krek, W. Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 2003, 302, 1208–1212. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Mihashi, K. Binding mode of cytochalasin B to F-actin is altered by lateral binding of regulatory proteins. J. Biochem. 1991, 109, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Shelanski, M.L.; Gaskin, F.; Cantor, C.R. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. USA 1973, 70, 765–768. [Google Scholar] [CrossRef] [PubMed]
- Ishii, K.; Noda, M.; Yagi, H.; Thammaporn, R.; Seetaha, S.; Satoh, T.; Kato, K.; Uchiyama, S. Disassembly of the self-assembled, double-ring structure of proteasome alpha7 homo-tetradecamer by alpha6. Sci. Rep. 2015, 5, 18167. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morita, K.; Yamamoto, Y.Y.; Hori, A.; Obata, T.; Uno, Y.; Shinohara, K.; Noguchi, K.; Noi, K.; Ogura, T.; Ishii, K.; et al. Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum. Int. J. Mol. Sci. 2018, 19, 2452. https://doi.org/10.3390/ijms19082452
Morita K, Yamamoto YY, Hori A, Obata T, Uno Y, Shinohara K, Noguchi K, Noi K, Ogura T, Ishii K, et al. Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum. International Journal of Molecular Sciences. 2018; 19(8):2452. https://doi.org/10.3390/ijms19082452
Chicago/Turabian StyleMorita, Kento, Yohei Y. Yamamoto, Ayaka Hori, Tomohiro Obata, Yuko Uno, Kyosuke Shinohara, Keiichi Noguchi, Kentaro Noi, Teru Ogura, Kentaro Ishii, and et al. 2018. "Expression, Functional Characterization, and Preliminary Crystallization of the Cochaperone Prefoldin from the Thermophilic Fungus Chaetomium thermophilum" International Journal of Molecular Sciences 19, no. 8: 2452. https://doi.org/10.3390/ijms19082452