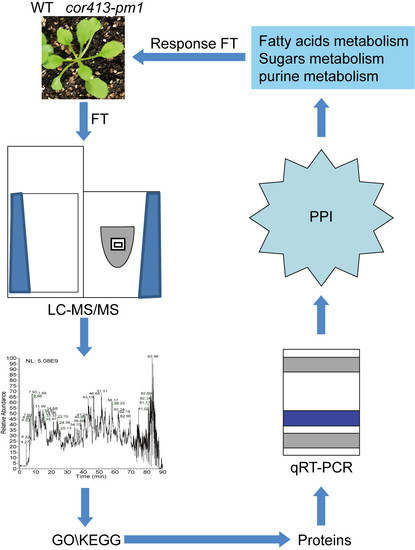
Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis
Abstract
:1. Introduction
2. Results
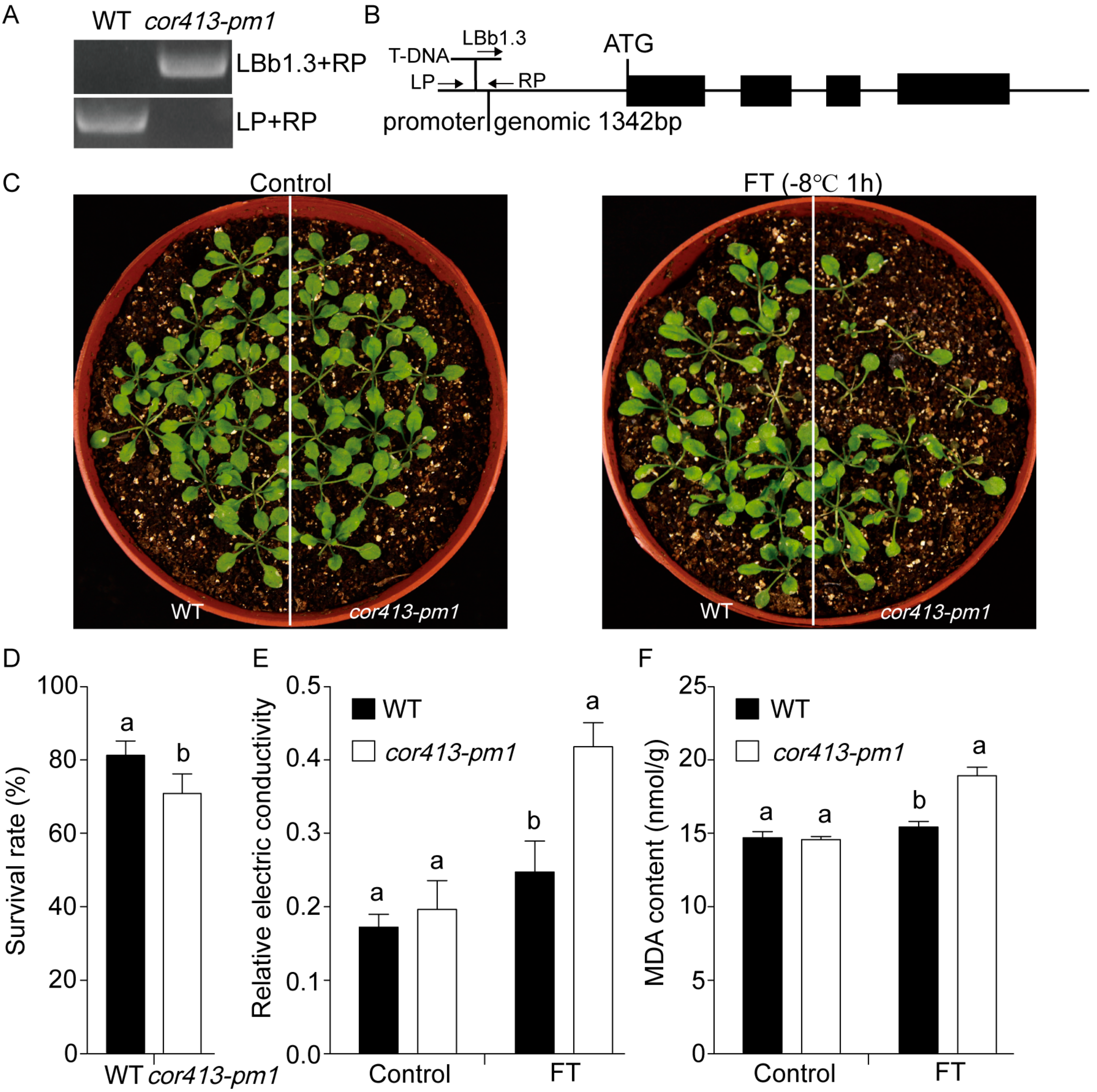
2.1. Gene Functions of COR413-PM1
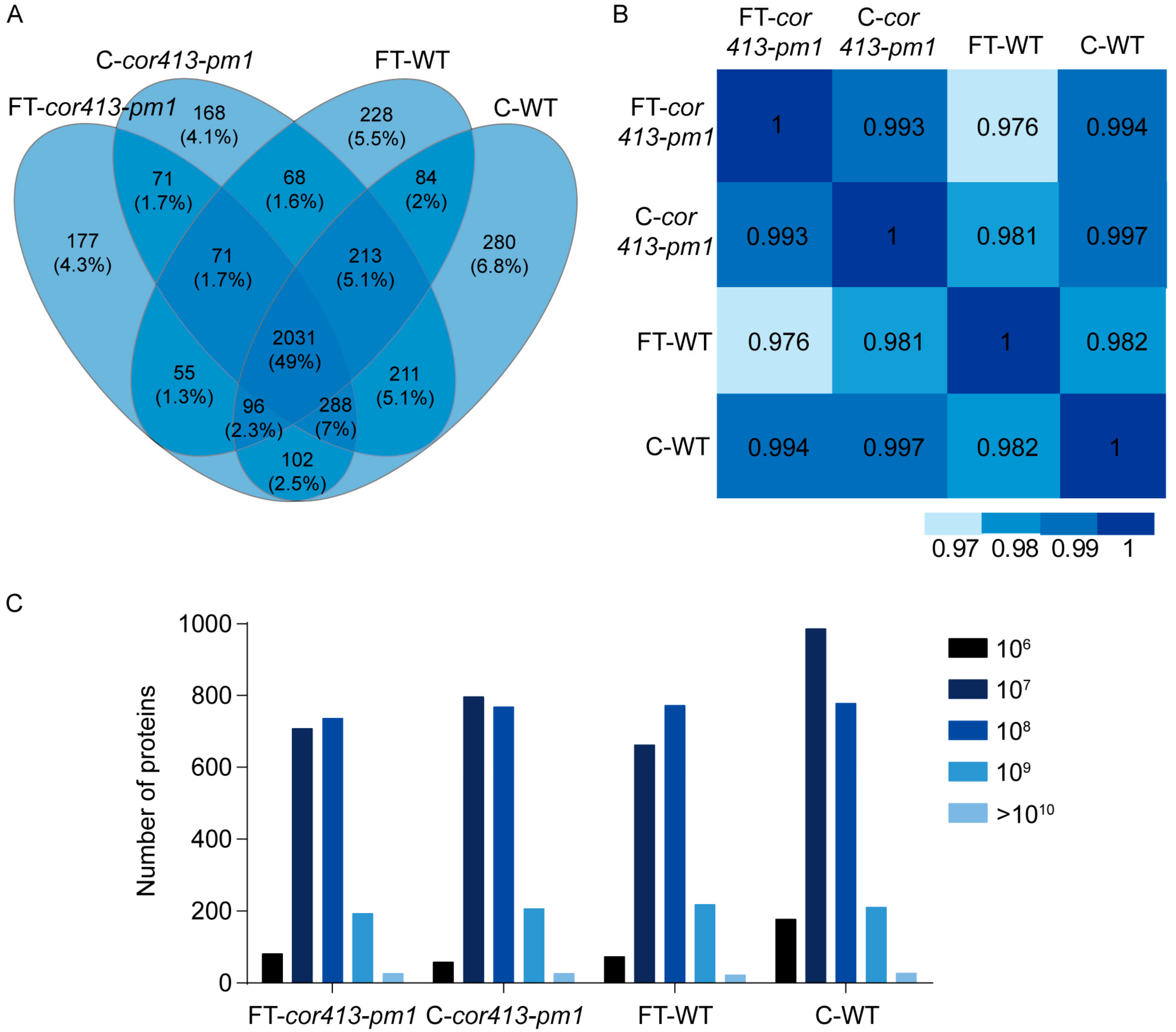
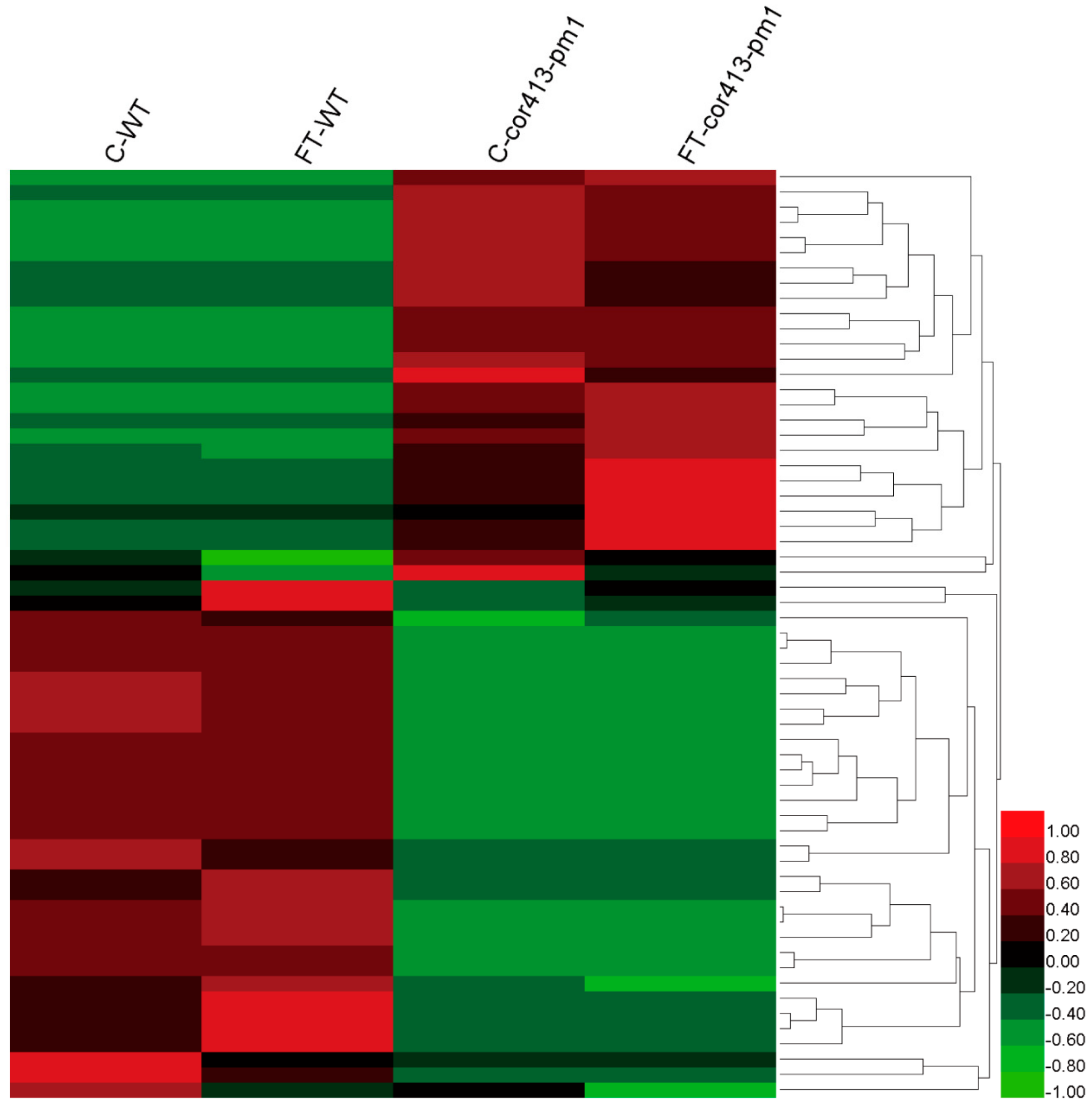
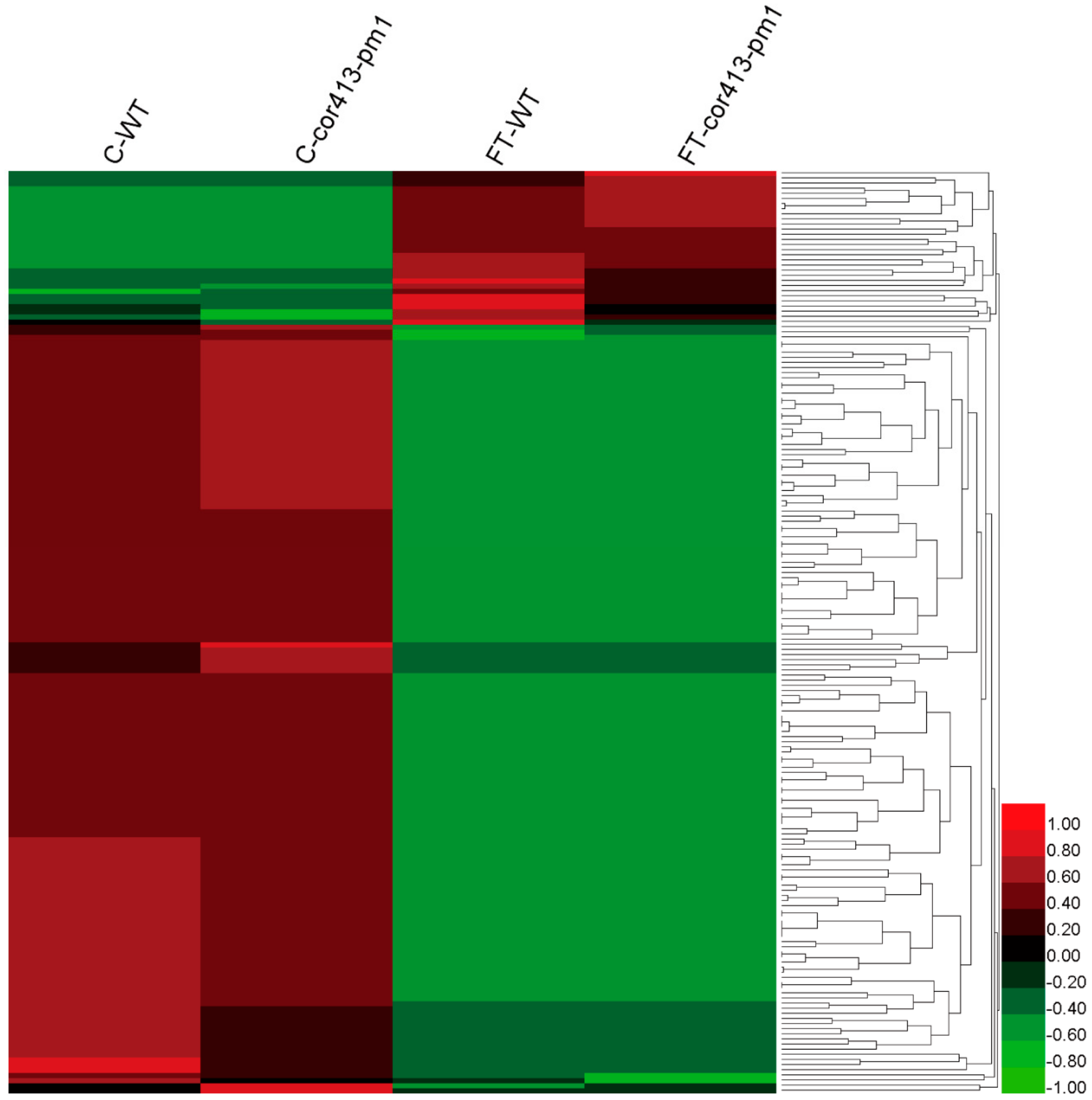
2.2. Identification of Differentially Abundant Proteins (DAPs) in Seedling Shoots of cor413-pm1 and WT
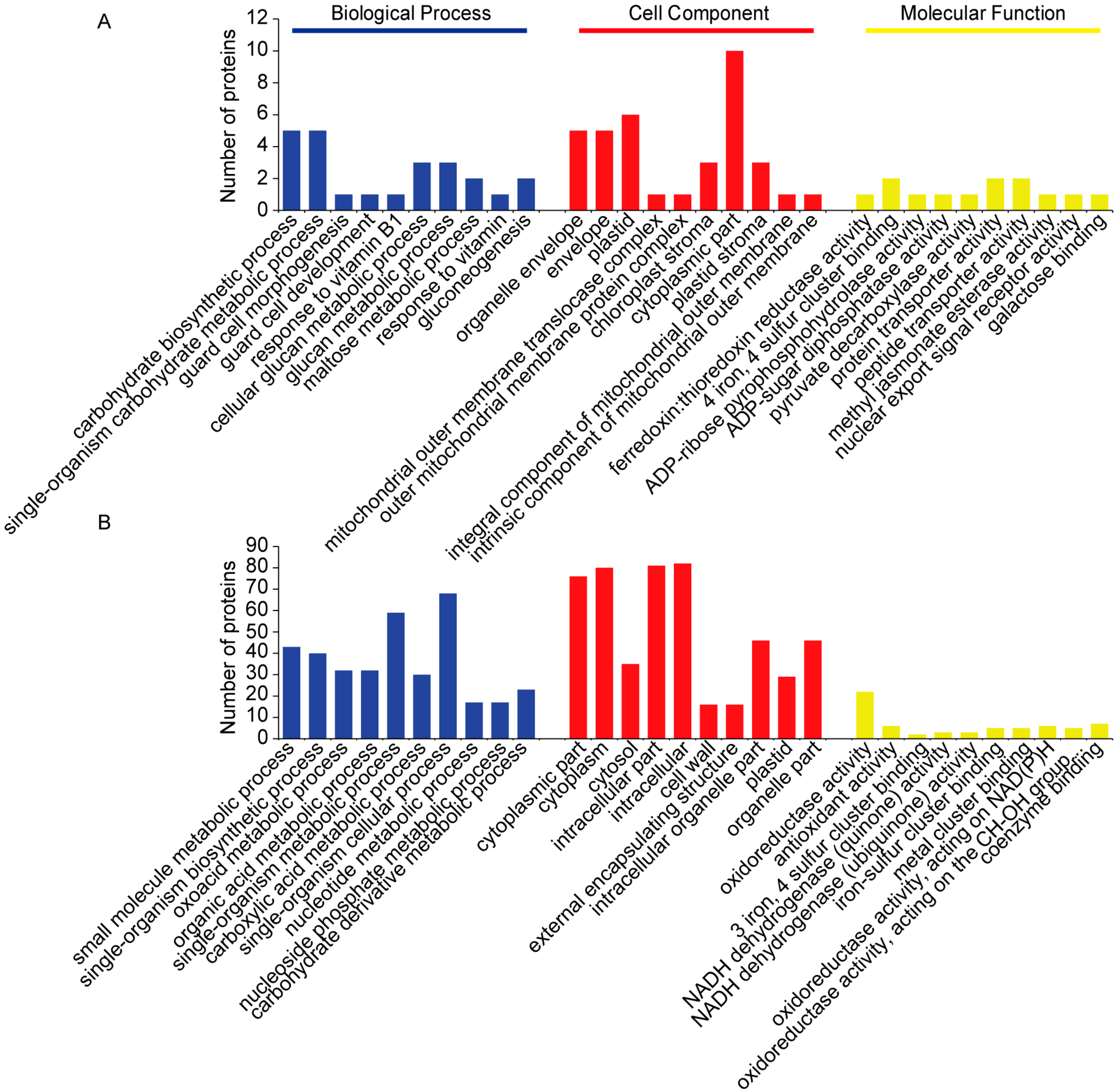
2.3. Categorization of DAPs in Seedling Shoots
2.4. qRT-PCR Analysis
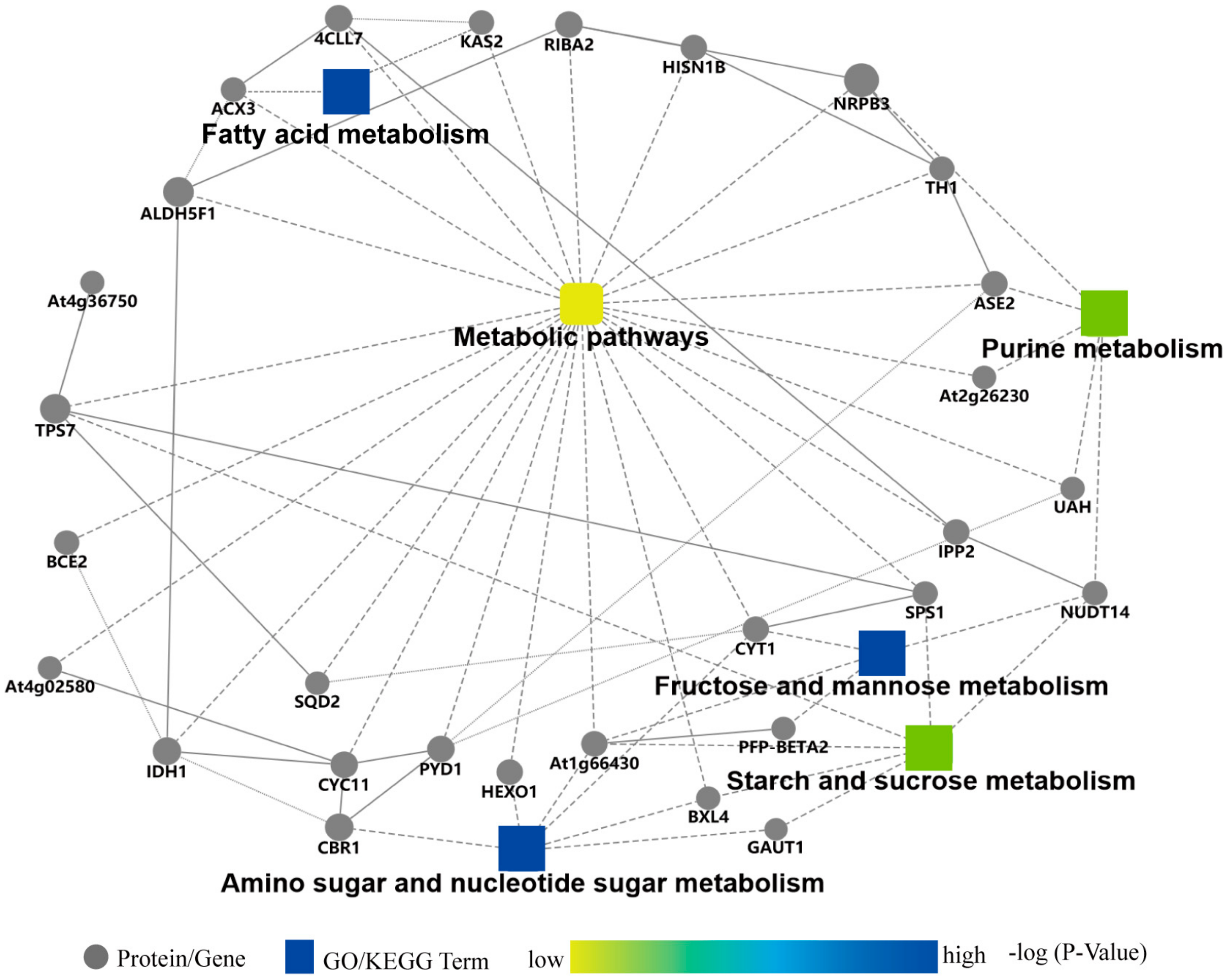
2.5. Protein–Protein Interaction Analysis of DAPs
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Relative Electrical Conductivity
4.3. Protein Extraction
4.4. Mass Spectrometric Analysis
4.5. Protein Identification and Quantitation
4.6. Bioinformatics Analysis
4.7. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
4.8. Accession Codes
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| COR | Cold-regulated |
| MDA | Malondialdehyde |
| MS | Mass Spectrometry |
| DAPs | Differentially Abundant Proteins |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | quantitative Real-Time Polymerase Chain Reaction |
References
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huo, W.; Zhang, L.; Chen, F.; Cui, D. Identification of winter-responsive proteins in bread wheat using proteomics analysis and virus-induced gene silencing. Mol. Cell. Proteom. 2016, 15, 2954–2969. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, C.; Zhu, J.-K. Molecular genetic analysis of cold-regulated gene transcription. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, M.F. Plant Cold Acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 571–599. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; Xin, Z. Temperature sensing and cold acclimation. Curr. Opin. Plant Biol. 2001, 4, 241–246. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeaty/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozakib, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Kosová, K.; Vítámvás, P.; Prášil, I.T.; Renaut, J. Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteom. 2011, 74, 1301–1322. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Hannah, M.A.; Wiese, D.; Freund, S.; Fiehn, O.; Heyer, A.G.; Hincha, D.K. Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol. 2006, 142, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Wanner, L.A.; Junttila, O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999, 120, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Xin, Z.; Browse, J. Cold comfort farm: The acclimation of plants to freezing temperatures. Plant Cell Environ. 2000, 23, 893–902. [Google Scholar] [CrossRef]
- Sung, D.-Y.; Kaplan, F.; Lee, K.-J.; Guy, C.L. Acquired tolerance to temperature extremes. Trends Plant Sci. 2003, 8, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.-H.; Henderson, D.A.; Zhu, J.-K. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 2005, 17, 3155–3175. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Buskirk, H.A.V.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Ishida, J.; Narusaka, M.; Fujita, M.; Nanjo, T.; Umezawa, T.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T. Monitoring the expression pattern of around 7000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct. Integr. Genom. 2002, 2, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature or high salt stress. Plant Cell 1994, 6, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, H.; Ren, F.; Guo, Q.-Q.; Hu, X.-P.; Li, X.-B. A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell Rep. 2011, 30, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Breton, G.; Danyluk, J.; Charron, J.-B.F.; Sarhan, F. Expression profiling and bioinformatic analyses of a novel stress-regulated multispanning transmembrane protein family from cereals and Arabidopsis. Plant Physiol. 2003, 132, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Machuka, J.; Bashiardes, S.; Ruben, E.; Spooner, K.; Cuming, A.; Knight, C.; Cove, D. Sequence analysis of expressed sequence tags from an ABA-treated cDNA 1ibrary identifies stress response genes in the moss Physcomitrella patens. Plant Cell Physiol. 1999, 40, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Orvar, B.L.; Sangwan, V.; Omann, F.; Dhindsa, R.S. Early steps in cold sensing by plant cells: The role of actin cytoskeleton and membrane fluidity. Plant J. 2000, 23, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Foulds, I.; Singh, J.; Dhindsa, R.S. Cold-activation of Brassica napus BN115 promoter is mediated by structural changes in membranes and cytoskeleton, and requires Ca2+ influx. Plant J. 2001, 27, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Thalhammer, A.; Hundertmark, M.; Popova, A.V.; Seckle, R.; Hincha, D.K. Interaction of two intrinsically disordered plant stress proteins (COR15A and COR15B) with lipid membranes in the dry state. Biochim. Biophys. Acta 2010, 1798, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Artus, N.N.; Uemura, M.; Steponkus, P.L.; Gilmour, S.J.; Lin, C.; Thomashow, M.F. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. USA 1996, 93, 13404–13409. [Google Scholar] [CrossRef] [PubMed]
- Gilmour, S.J.; Artus, N.N.; Thomashow, M.F. cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 1992, 18, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Danyluk, J.; Perron, A.; Houde, M.; Limin, A.; Fowler, B.; Benhamou, N.; Sarhan, F. Accumulation of an acidic dehydrin in the vicinity of the plasma membrane during cold acclimation of wheat. Plant Cell 1998, 10, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Janmohammadi, M.; Zolla, L.; Rinalducci, S. Low temperature tolerance in plants: Changes at the protein level. Phytochemistry 2015, 117, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Bouché, N.; Bouchez, D. Arabidopsis gene knockout: Phenotypes wanted. Curr. Opin. Plant Biol. 2001, 4, 111–117. [Google Scholar] [CrossRef]
- Parinov, S.; Sundaresan, V. Functional genomics in Arabidopsis: Large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol. 2000, 11, 157–161. [Google Scholar] [CrossRef]
- Jiang, B.; Shi, Y.; Zhang, X.; Xin, X.; Qi, L.; Guo, H.; Li, J.; Yang, S. PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E6695–E6702. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, D.; Li, C.; Hao, Y.; Ma, F.; Shu, H. Influence of drought stress on the cellular ultrastructure and antioxidant system in leaves of drought-tolerant and drought-sensitive apple rootstocks. Plant Physiol. Biochem. 2012, 51, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bogeat-Triboulot, M.-B.; Brosche, M.; Renaut, J.; Jouve, L.; Thiec, D.L.; Fayyaz, P.; Vinocur, B.; Witters, E.; Laukens, K.; Teichmann, T. Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiol. 2007, 143, 876–892. [Google Scholar] [CrossRef] [PubMed]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef] [PubMed]
- Renauta, J.; Hausman, J.-F.; Wisniewski, M.E. Proteomics and low-temperature studies: Bridging the gap between gene expression and metabolism. Physiol. Plant. 2006, 126, 97–109. [Google Scholar] [CrossRef]
- Abdelmegid, S.; Murugaiyan, J.; Abo-Ismail, M.; Caswell, J.L.; Kelton, D.; Kirby, G.M. Identification of host defense-related proteins using label-free quantitative proteomic analysis of milk whey from cows with staphylococcus aureus subclinical mastitis. Int. J. Mol. Sci. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Maki, K.; Takeshi, O.; Bunichi, E. Salt stress-induced lipid peroxidation is reduced by glutathione S-transferase, but this reduction of lipid peroxides is not enough for a recovery of root growth in Arabidopsis. Plant Sci. 2005, 169, 369–373. [Google Scholar] [CrossRef]
- Mohapatra, S.S.; Poole, R.J.; Dhindsa, R.S. Alterations in membrane protein-profile during cold treatment of Alfalfa. Plant Physiol. 1988, 86, 1005–1007. [Google Scholar] [CrossRef] [PubMed]
- Imai, R.; Koike, M.; Sutoh, K.; Kawakami, A.; Torada, A.; Oono, K. Molecular characterization of a cold-induced plasma membrane protein gene from wheat. Mol. Genet. Genom. 2005, 274, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Uemura, M.; Steponkus, P.L. Cold acclimation in plants: Relationship between the lipid composition and the cryostability of the plasma membrane. J. Plant Res. 1999, 112, 245–254. [Google Scholar] [CrossRef]
- Lyons, J.M.; Asmundson, C.M. Solidification of unsaturated/saturated fatty acid mixtures and its relationship to chilling sensitivity in plants. J. Am. Oil Chem. Soc. 1965, 42, 1056–1058. [Google Scholar] [CrossRef] [PubMed]
- Kodama, H.; Horiguchi, C.; Nishiuchi, T.; Nishimura, M.; Iba, K. Fatty acid desaturation during chilling acclimation is one of the factors involved in conferring low-temperature tolerance to young tobacco leaves. Plant Physiol. 1995, 107, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Murata, N.; Ishizaki-Nishizawa, O.; Higashi, S.; Hayashi, H.; Tasaka, Y.; Nishida, I. Genetically engineered alteration in the chilling sensitivity of plants. Nature 1992, 356, 710–713. [Google Scholar] [CrossRef]
- Yehuda, S.; Rabinovitz, S.; Carasso, R.L.; Mostofsky, D.I. The role of polyunsaturated fatty acids in restoring the aging neuronal membrane. Neurobiol. Aging 2002, 23, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Lightner, J.; Warwick, N.; Browse, J. Low-temperature damage and subsequent recovery of fab1 mutant Arabidopsis exposed to 2 °C. Plant Physiol. 1997, 113, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, A.S.; LaBrie, S.T.; Kinney, A.J.; Wettstein-Knowles, P.; Browse, J. A KAS2 cDNA complements the phenotypes of the Arabidopsis fab1 mutant that differs in a single residue bordering the substrate binding pocket. Plant J. 2002, 29, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; James, D.W., Jr.; Dooner, H.K.; Browse, J. A mutant of Arabidopsis deficient in the elongation of palmitic acid. Plant Physiol. 1994, 106, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wallis, J.G.; Browse, J. Mutations in the prokaryotic pathway rescue the fatty acid biosynthesis 1 mutant in the cold. Plant Physiol. 2015, 169, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Sturm, A. Invertases. primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999, 121, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Riggs, J.W.; Cavales, P.C.; Chapiro, S.M.; Callis, J. Identification and biochemical characterization of the fructokinase gene family in Arabidopsis thaliana. BMC Plant Biol. 2017, 17, 83. [Google Scholar] [CrossRef] [PubMed]
- Stein, O.; Avin-Wittenberg, T.; Krahnert, I.; Zemach, H.; Bogol, V.; Daron, O.; Aloni, R.; Fernie, A.R.; Granot, D. Arabidopsis fructokinases are important for seed oil accumulation and vascular development. Front. Plant Sci. 2017, 7, 2047. [Google Scholar] [CrossRef] [PubMed]
- Roach, M.; Gerber, L.; Sandquist, D.; Gorzsas, A.; Hedenstrom, M.; Kumar, M.; Steinhauser, M.C.; Feil, R.; Daniel, G.; Stitt, M. Fructokinase is required for carbon partitioning to cellulose in aspen wood. Plant J. 2012, 70, 967–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Liua, L. Expression and crystallographic studies of the Arabidopsis thaliana GDP-D-mannose pyrophosphorylase VTC1. Acta Crystallogr. F Struct Biol. Commun. 2016, 72, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Zhang, R.; Huang, R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J. 2012, 71, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Li, Z. Transcriptome profile and differentially expressed genes analysis in winter wheat under cold stress conditions. Res. J. Biotechnol. 2015, 10, 73–88. [Google Scholar]
- Strand, A.; Foyer, C.H.; Gustafsson, P.; Gardestrom, P.; Hurry, V. Altering flux through the sucrose biosynthesis pathway in transgenic Arabidopsis thaliana modifies photosynthetic acclimation at low temperatures and the development of freezing tolerance. Plant Cell Environ. 2003, 26, 523–535. [Google Scholar] [CrossRef]
- Stasolla, C.; Katahira, R.; Thorpe, T.A.; Ashihara, H. Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 2003, 160, 1271–1295. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.F.; Chen, L.J.; Boldt, R.; Sun, C.W.; Li, H.M.; Hung, W.-F.; Chen, L.-J.; Boldt, R.; Sun, C.-W.; Li, H.-M. Characterization of Arabidopsis glutamine phosphoribosyl pyrophosphate amidotransferase-deficient mutants. Plant Physiol. 2004, 135, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Rapacz, M.; Wolanin, B.; Hura, K.; Tyrka, M. The effects of cold acclimation on photosynthetic apparatus and the expression of COR14b in four genotypes of barley (Hordeum vulgare) contrasting in their tolerance to freezing and high-light treatment in cold conditions. Ann. Bot. 2008, 101, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Ensminger, I.; Busch, F.; Huner, N.P.A. Photostasis and cold acclimation: Sensing low temperature through photosynthesis. Physiol. Plant. 2006, 126, 28–44. [Google Scholar] [CrossRef]
- Van der Graaff, E.; Hooykaas, P.; Lein, W.; Lerchl, J.; Kunze, G.; Sonnewald, U.; Boldt, R. Molecular analysis of “de novo” purine biosynthesis in solanaceous species and in Arabidopsis thaliana. Front. Biosci. 2004, 9, 1803–1816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, S.; Zhang, Y.; Wang, X.; Li, D.; Li, Q.; Yue, M.; Li, Q.; Zhang, Y.-E.; Xu, Y. Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 2011, 23, 396–411. [Google Scholar] [CrossRef] [PubMed]
- Cornelius, S.; Witz, S.; Rolletschek, H.; Mohlmann, T. Pyrimidine degradation influences germination seedling growth and production of Arabidopsis seeds. J. Exp. Bot. 2011, 62, 5623–5632. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]
- Vizcaino, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
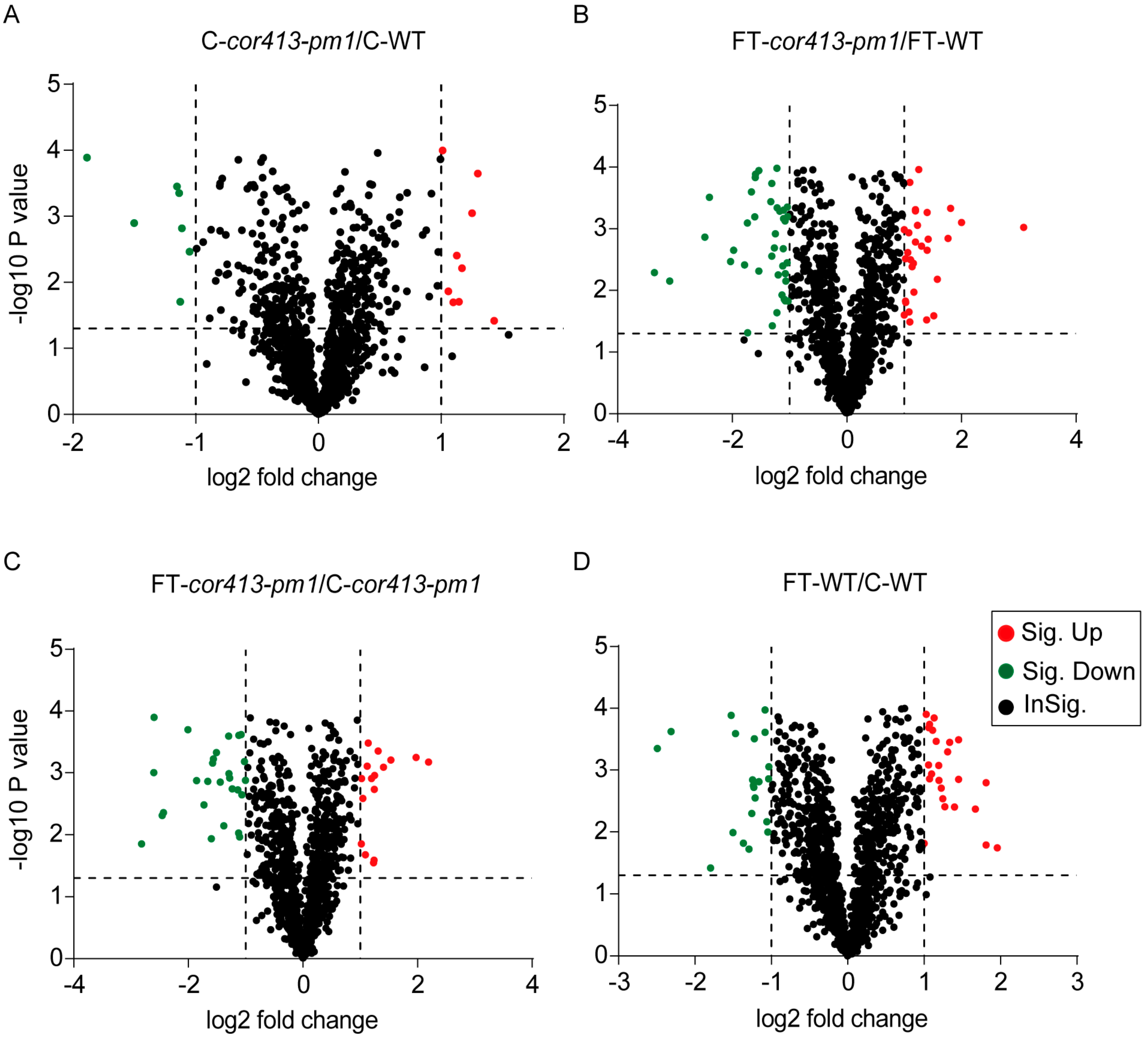




| Treatment Groups | Upregulated Proteins | Downregulated Proteins | Unchanged Proteins | Total Proteins |
|---|---|---|---|---|
| FT-cor413-pm1 1 vs. FT-WT 2 | 552 (a) | 573 (c) | 1125 | 2250 |
| C-cor413-pm1 3 vs. C-WT 4 | 298 (b) | 625 (d) | 1537 | 2460 |
| FT-cor413-pm1 vs. C-cor413-pm1 | 407 (e) | 539 (g) | 1288 | 2234 |
| FT-WT vs. C-WT | 458 (f) | 888 (h) | 1243 | 2589 |
| Name | Resolution Ratio | AGC Target | Maximum IT 1 | Scan Range | Top N | NCE |
|---|---|---|---|---|---|---|
| Full MS | 70,000 | 3 × 106 | 50 ms | 300–1800 m/z | / | / |
| dd-MS2 | 17,500 | 1 × 105 | 45 ms | / | 20 | 27 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, C.; Chen, K.; Ding, Q.; Mou, Y.; Yang, R.; Zhao, M.; Ma, B.; Xu, Z.; Ma, Y.; Pan, Y.; et al. Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2572. https://doi.org/10.3390/ijms19092572
Su C, Chen K, Ding Q, Mou Y, Yang R, Zhao M, Ma B, Xu Z, Ma Y, Pan Y, et al. Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(9):2572. https://doi.org/10.3390/ijms19092572
Chicago/Turabian StyleSu, Chen, Kai Chen, Qingqian Ding, Yongying Mou, Rui Yang, Mengjie Zhao, Bo Ma, Zhaoshi Xu, Youzhi Ma, Yinghong Pan, and et al. 2018. "Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis" International Journal of Molecular Sciences 19, no. 9: 2572. https://doi.org/10.3390/ijms19092572
APA StyleSu, C., Chen, K., Ding, Q., Mou, Y., Yang, R., Zhao, M., Ma, B., Xu, Z., Ma, Y., Pan, Y., Chen, M., & Xi, Y. (2018). Proteomic Analysis of the Function of a Novel Cold-Regulated Multispanning Transmembrane Protein COR413-PM1 in Arabidopsis. International Journal of Molecular Sciences, 19(9), 2572. https://doi.org/10.3390/ijms19092572