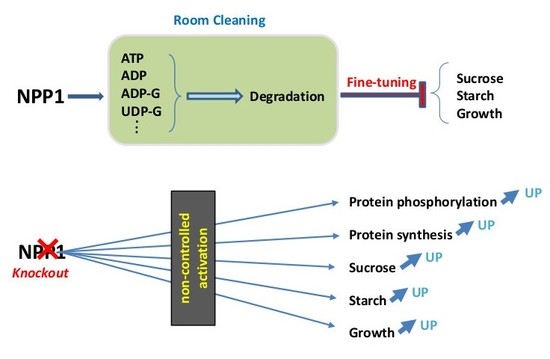
Proteomics Analysis Reveals Non-Controlled Activation of Photosynthesis and Protein Synthesis in a Rice npp1 Mutant under High Temperature and Elevated CO2 Conditions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Knocking out NPP1 Decreases Leaf Temperature and Enhances Stomatal Conductance under High Temperature and Elevated CO2 Conditions
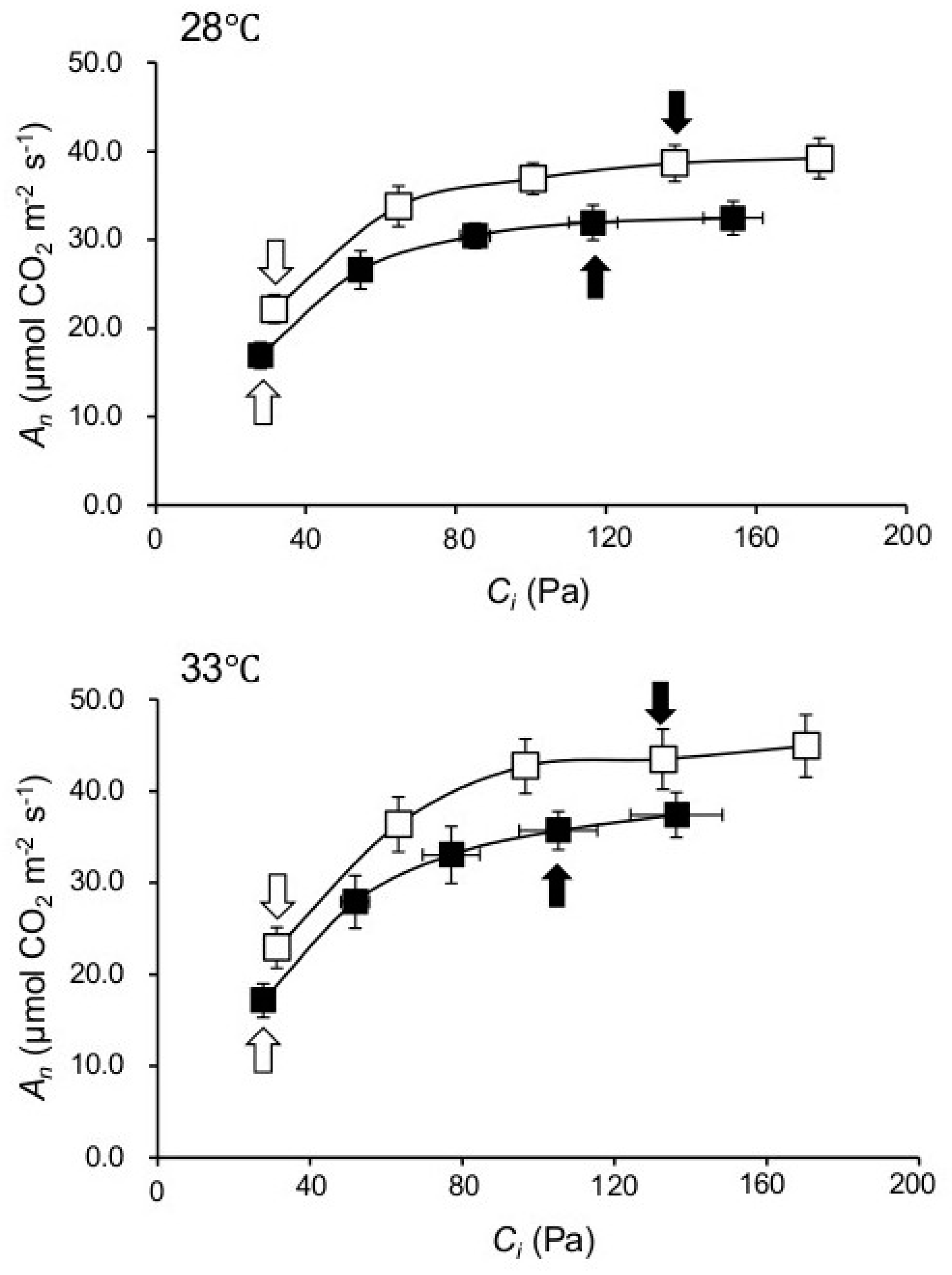
2.2. Knocking out NPP1 Enhances Photosynthetic Capacity under High Temperature and Elevated CO2 Conditions
2.3. Proteomic Characterization of npp1 Leaves under High Temperature and Elevated CO2 Conditions
2.4. Photosynthesis and Carbohydrate Metabolism
2.5. Protein Synthesis System
2.6. Signaling
3. Material and Methods
3.1. Plant Material and Growth Condition
3.2. Thermal Imaging
3.3. Gas Exchange Measurements
3.4. Assay
3.5. Analysis of Leaf Proteome
3.6. Analysis of Chloroplast Phosphoproteome
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ray, D.K.; Gerber, J.S.; MacDonald, G.K.; West, P.C. Climate variation explains a third of global crop yield variability. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Becklin, K.M.; Anderson, J.T.; Gerhart, L.M.; Wadgymar, S.M.; Wessinger, C.A.; Ward, J.K. Examining plant physiological responses to climate change through an evolutionary Lens. Plant Physiol. 2016, 172, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Becklin, K.M.; Walker, S.M.; Way, D.A.; Ward, J.K. CO2 studies remain key to understanding a future world. New Phytol. 2017, 214, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; IIda, Y.; Ishihara, K. Effect of leaf water potential and air humidity on photosynthetic rate and diffusive conductance in rice plants. Jpn. J. Crop Sci. 1988, 57, 112–118. [Google Scholar] [CrossRef]
- Ishihara, K.; Saitoh, K. Diurnal courses of photosynthesis, transpiration, and diffusive conductance in the single-leaf of the rice plants grown in the paddy field under submerged condition. Jpn. J. Crop Sci. 1987, 56, 8–17. [Google Scholar] [CrossRef]
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Kusumi, K.; Chono, Y.; Shimada, H.; Gotoh, E.; Tsuyama, M.; Iba, K. Chloroplast biogenesis during the early stage of leaf development in rice. Plant Biotechnol. 2010, 27, 85–90. [Google Scholar] [CrossRef] [Green Version]
- Ishimaru, K. Identification of a locus increasing rice yield and physiological analysis of its function. Plant Physiol. 2003, 133, 1083–1090. [Google Scholar] [CrossRef] [PubMed]
- Scofield, G.N.; Hirose, T.; Aoki, N.; Furbank, R.T. Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J. Exp. Bot. 2007, 58, 3155–3169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahaji, A.; Li, J.; Sánchez-López, Á.M.; Baroja-Fernández, E.; Muñoz, F.J.; Ovecka, M.; Almagro, G.; Montero, M.; Ezquer, I.; Etxeberria, E.; et al. Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 2014, 32, 87–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ball, S.G.; Morell, M.K. From bacterial glycogen to starch: Understanding the Biogenesis of the Plant Starch Granule. Annu. Rev. Plant Biol. 2003, 54, 207–233. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-López, M.; Baroja-Fernández, E.; Zandueta-Criado, A.; Pozueta-Romero, J. Adenosine diphosphate glucose pyrophosphatase: A plastidial phosphodiesterase that prevents starch biosynthesis. Proc. Natl. Acad. Sci. USA 2000, 97, 8705–8710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanjo, Y.; Oka, H.; Ikarashi, N.; Kaneko, K.; Kitajima, A.; Mitsui, T.; Muñoz, F.J.; Rodríguez-López, M.; Baroja-Fernández, E.; Pozueta-Romero, J. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-golgi to the chloroplast through the secretory pathway. Plant Cell 2006, 18, 2582–2592. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Inomata, T.; Masui, T.; Koshu, T.; Umezawa, Y.; Itoh, K.; Pozueta-romero, J.; Mitsui, T. Nucleotide Pyrophosphatase/Phosphodiesterase 1 exerts a negative effect on starch accumulation and growth in rice seedlings under high temperature and CO2 concentration conditions. Plant Cell Physiol. 2014, 55, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Yamada, C.; Yanagida, A.; Koshu, T.; Umezawa, Y.; Itoh, K.; Hori, H.; Mitsui, T. Differential localizations and functions of rice nucleotide pyrophosphatase/phosphodiesterase isozymes 1 and 3. Plant Biotechnol. 2011, 28, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Kaneko, K.; Takamatsu, T.; Inomata, T.; Oikawa, K.; Itoh, K.; Hirose, K.; Amano, M.; Nishimura, S.-I.; Toyooka, K.; Matsuoka, K.; et al. N-glycomic and microscopic subcellular localization analyses of NPP1, 2 and 6 strongly indicate that trans-Golgi compartments participate in the Golgi-to-plastid traffic of nucleotide pyrophosphatase/phosphodiesterases in rice. Plant Cell Physiol. 2016, 57, 1610–1628. [Google Scholar] [CrossRef] [PubMed]
- Baslam, M.; Oikawa, K.; Kitajima-koga, A.; Kaneko, K.; Mitsui, T. Golgi-to-plastid trafficking of proteins through secretory pathway: Insights into vesicle-mediated import toward the plastids. Plant Signal. Behav. 2016, 11, e1221558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asatsuma, S.; Sawada, C.; Itoh, K.; Okito, M.; Kitajima, A.; Mitsui, T. Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005, 46, 858–869. [Google Scholar] [CrossRef] [PubMed]
- Villarejo, A.; Burén, S.; Larsson, S.; Déjardin, A.; Monné, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, A.; Asatsuma, S.; Okada, H.; Hamada, Y.; Kaneko, K.; Nanjo, Y.; Kawagoe, Y.; Toyooka, K.; Matsuoka, K.; Takeuchi, M.; et al. The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 2009, 21, 2844–2858. [Google Scholar] [CrossRef] [PubMed]
- Burén, S.; Ortega-Villasante, C.; Blanco-Rivero, A.; Martínez-Bernardini, A.; Shutova, T.; Shevela, D.; Messinger, J.; Bako, L.; Villarejo, A.; Samuelsson, G. Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS ONE 2011, 6, e21021. [Google Scholar] [CrossRef] [PubMed]
- Shiraya, T.; Mori, T.; Maruyama, T.; Sasaki, M.; Takamatsu, T.; Oikawa, K.; Itoh, K.; Kaneko, K.; Ichikawa, H.; Mitsui, T. Golgi/plastid-type manganese superoxide dismutase involved in heat-stress tolerance during grain filling of rice. Plant Biotechnol. J. 2015, 13, 1251–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.S.; Frank, J.; Kang, C.H.; Kajiura, H.; Vikram, M.; Ueda, A.; Kim, S.; Bahk, J.D.; Triplett, B.; Fujiyama, K.; et al. Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 2008, 105, 5933–5938. [Google Scholar] [CrossRef] [PubMed]
- Teskey, R.O.; Fites, J.A.; Samuelson, L.J.; Bongarten, B.C. Stomatal and nonstomatal limitations to net photosynthesis in Pinus taeda L. under different environmental conditions. Tree Physiol. 1986, 2, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F.; Sharkey, T.D. The effect of temperature on the occurrence of O2 and CO2 insensitive photosynthesis in field grown plants. Plant Physiol. 1987, 84, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Cerasoli, S.; Wertin, T.; McGuire, M.A.; Rodrigues, A.; Aubrey, D.P.; Pereira, J.S.; Teskey, R.O. Poplar saplings exposed to recurring temperature shifts of different amplitude exhibit differences in leaf gas exchange and growth despite equal mean temperature. AoB Plants 2014, 6, plu018. [Google Scholar] [CrossRef] [PubMed]
- Von Caemmerer, S.; Evans, J.R. Temperature responses of mesophyll conductance differ greatly between species. Plant. Cell Environ. 2015, 38, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Uprety, D.C.; Dwivedi, N.; Jain, V.; Mohan, R. Effect of elevated carbon dioxide concentration on the stomatal parameters of rice cultivars. Photosynthetica 2002, 40, 315–319. [Google Scholar] [CrossRef]
- Shimazaki, K.; Iino, M.; Zeiger, E. Blue light-dependent proton extrusion by guard-cell protoplasts of Vicia faba. Nature 1986, 319, 324–326. [Google Scholar] [CrossRef]
- Talbott, L.D.; Zeiger, E. The role of sucrose in guard cell osmoregulation. J. Exp. Bot. 1998, 49, 329–337. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Hagiwara, S. Cytosolic calcium regulates ion channels in the plasma membrane of Vicia faba guard cells. Nature 1989, 338, 427–430. [Google Scholar] [CrossRef]
- Kusumi, K.; Hirotsuka, S.; Kumamaru, T.; Iba, K. Increased leaf photosynthesis caused by elevated stomatal conductance in a rice mutant deficient in SLAC1, a guard cell anion channel protein. J. Exp. Bot. 2012, 63, 5635–5644. [Google Scholar] [CrossRef] [PubMed]
- Portis, A. Rubisco Activase—Rubisco’s catalytic chaperone. Photosynth. Res. 2003, 75, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Baroja-Fernandez, E.; Bahaji, A.; Munoz, F.J.; Ovecka, M.; Montero, M.; Sesma, M.T.; Alonso-Casajus, N.; Almagro, G.; Sanchez-Lopez, A.M.; et al. Enhancing sucrose synthase activity results in increased levels of starch and ADP-glucose in maize (Zea mays L.) seed endosperms. Plant Cell Physiol. 2013, 54, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gehan, J.P.; Sharkey, T.D. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005, 138, 2280–2291. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chang, L.; Tong, Z.; Wang, D.; Yin, Q.; Wang, D.; Jin, X.; Yang, Q.; Wang, L.; Sun, Y.; et al. Proteomics profiling reveals carbohydrate metabolic enzymes and 14-3-3 proteins play important roles for starch accumulation during cassava root tuberization. Sci. Rep. 2016, 6, 19643. [Google Scholar] [CrossRef] [PubMed]
- Clore, A.M.; Dannenhoffer, J.M.; Larkins, B.A. EF-1α is associated with a cytoskeletal network surrounding protein bodies in maize endosperm cells. Plant Cell 1996, 8, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Yahalom, A.; Kim, T.-H.; Roy, B.; Singer, R.; Von Arnim, A.G.; Chamovitz, D.A. Arabidopsis eIF3e is regulated by the COP9 signalosome and has an impact on development and protein translation. Plant J. 2007, 53, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Deng, Z.; Gong, C.; Wang, T. The rice eukaryotic translation initiation factor 3 subunit f (OseIF3f) is involved in microgametogenesis. Front. Plant Sci. 2016, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-H.; Kim, B.-H.; Yahalom, A.; Chamovitz, D.A.; von Arnim, A.G. Translational regulation via 5’ mRNA leader sequences revealed by mutational analysis of the Arabidopsis translation initiation factor subunit eIF3h. Plant Cell 2004, 16, 3341–3356. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Subramanian, R.R.; Masters, S.C. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000, 40, 617–647. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Salinas, J.; Collinge, D.B. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 2002, 50, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.S.; Swatek, K.N.; Thelen, J.J. Regulation of the regulators: Post-Translational Modifications, subcellular, and spatiotemporal distribution of plant 14-3-3 proteins. Front. Plant Sci. 2016, 7, 611. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Jia, L.; Shi, W.; Liang, J.; Zhang, J. Smart role of plant 14-3-3 proteins in response to phosphate deficiency. Plant Signal. Behav. 2012, 7, 1047–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sehnke, P.C.; Chung, H.J.; Wu, K.; Ferl, R.J. Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 765–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baunsgaard, L.; Fuglsang, A.T.; Jahn, T.; Korthout, H.A.; de Boer, A.H.; Palmgren, M.G. The 14-3-3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998, 13, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Cotelle, V.; Meek, S.E.; Provan, F.; Milne, F.C.; Morrice, N.; MacKintosh, C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 2000, 19, 2869–2876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moorhead, G.; Douglas, P.; Cotelle, V.; Harthill, J.; Morrice, N.; Meek, S.; Deiting, U.; Stitt, M.; Scarabel, M.; Aitken, A.; et al. Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 1999, 18, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toroser, D.; Athwal, G.S.; Huber, S.C. Site-specific regulatory interaction between spinach leaf sucrose-phosphate synthase and 14-3-3 proteins. FEBS Lett. 1998, 435, 110–114. [Google Scholar] [CrossRef] [Green Version]
- Diaz, C.; Kusano, M.; Sulpice, R.; Araki, M.; Redestig, H.; Saito, K.; Stitt, M.; Shin, R. Determining novel functions of Arabidopsis 14-3-3 proteins in central metabolic processes. BMC Syst. Biol. 2011, 5, 192. [Google Scholar] [CrossRef] [PubMed]
- Sehnke, P.C.; Henry, R.; Cline, K.; Ferl, R.J. Interaction of a plant 14-3-3 protein with the signal peptide of a thylakoid-targeted chloroplast precursor protein and the presence of 14-3-3 isoforms in the chloroplast stroma. Plant Physiol. 2000, 122, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Bellafiore, S.; Barneche, F.; Peltier, G.; Rochaix, J.-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 2005, 433, 892–895. [Google Scholar] [CrossRef] [PubMed]
- Bonardi, V.; Pesaresi, P.; Becker, T.; Schleiff, E.; Wagner, R.; Pfannschmidt, T.; Jahns, P.; Leister, D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 2005, 437, 1179–1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrzykowska, M.; Suorsa, M.; Semchonok, D.A.; Tikkanen, M.; Boekema, E.J.; Aro, E.-M.; Jansson, S. The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 2014, 26, 3646–3660. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, W.; Zhang, B.; Zhou, J.; Wang, Y.; Yang, Q.; Ke, Y.; He, H. Phosphoproteins regulated by heat stress in rice leaves. Proteome Sci. 2011, 9, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemeille, S.; Turkina, M.V.; Vener, A.V.; Rochaix, J.-D. Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteom. 2010, 9, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Facette, M.R.; Shen, Z.; Björnsdóttir, F.R.; Briggs, S.P.; Smith, L.G. Parallel proteomic and phosphoproteomic analyses of successive stages of maize leaf development. Plant Cell 2013, 25, 2798–2812. [Google Scholar] [CrossRef] [PubMed]
- Baginsky, S. Protein phosphorylation in chloroplasts—A survey of phosphorylation targets. J. Exp. Bot. 2016, 67, 3873–3882. [Google Scholar] [CrossRef] [PubMed]
- Rocha, A.G.; Mehlmer, N.; Stael, S.; Mair, A.; Parvin, N.; Chigri, F.; Teige, M.; Vothknecht, U.C. Phosphorylation of Arabidopsis transketolase at Ser 428 provides a potential paradigm for the metabolic control of chloroplast carbon metabolism. Biochem. J. 2014, 458, 313–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, M.; Kato, H.; Ogawa, T.; Kurachi, A.; Nakagawa, Y.; Kobayashi, H. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl. Acad. Sci. USA 2010, 107, 10760–10764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas-Suárez, M.; Castro-Sánchez, A.; Toledo-Ortiz, G.; González de la Vara, L.E.; García, E.; Loza-Tavera, H. Protein phosphorylation regulates in vitro spinach chloroplast petD mRNA 3′-untranslated region stability, processing, and degradation. Biochimie 2013, 95, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Tikkanen, M.; Aro, E.-M. Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci. 2014, 19, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, N.; Higa, T.; Kong, S.-G.; Wada, M. Plastid Movement Impaired1 and Plastid Movement Impaired1-Related1 mediate photorelocation movements of both chloroplasts and nuclei. Plant Physiol. 2015, 169, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.-J.; Oelmüller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.-P.; Chen, G.-X.; Yang, Z.-N. Regulatory role of Arabidopsis pTAC14 in chloroplast development and plastid gene expression. Plant Signal. Behav. 2012, 7, 1354–1356. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.-P.; Yu, Q.-B.; Zhao, T.-T.; Ma, Q.; Chen, G.-X.; Yang, Z.-N. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 2011, 157, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Miyao, A.; Tanaka, K.; Murata, K.; Sawaki, H.; Takeda, S.; Abe, K.; Shinozuka, Y.; Onosato, K.; Hirochika, H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 2003, 15, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, K.; Sasaki, M.; Kuribayashi, N.; Suzuki, H.; Sasuga, Y.; Shiraya, T.; Inomata, T.; Itoh, K.; Baslam, M.; Mitsui, T. Proteomic and glycomic characterization of rice chalky grains produced under moderate and high-temperature conditions in field system. Rice 2016, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Dorfer, V.; Pichler, P.; Stranzl, T.; Stadlmann, J.; Taus, T.; Winkler, S.; Mechtler, K. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. J. Proteome Res. 2014, 13, 3679–3684. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, T.; Baslam, M.; Inomata, T.; Oikawa, K.; Itoh, K.; Ohnishi, T.; Kinoshita, T.; Mitsui, T. Optimized method of extracting rice chloroplast DNA for high-quality plastome resequencing and de novo assembly. Front. Plant Sci. 2018, 9, 266. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, I.; Hirabayashi-Ishioka, Y.; Sakikawa, I.; Ota, T.; Yokoyama, M.; Uchiumi, T.; Morita, A. Optimization of enrichment conditions on TiO2 chromatography using glycerol as an additive reagent for effective phosphoproteomic analysis. J. Proteome Res. 2013, 12, 5587–5597. [Google Scholar] [CrossRef] [PubMed]







| Description | Accession | WT Control | WT HT&ECO2 | npp1 Control | npp1 HT&ECO2 |
|---|---|---|---|---|---|
| Chlorophyll a/b-binding protein | Q6Z411 | (1.15 ± 0.78) × 108 | (2.43 ± 2.06) × 108 | (1.05 ± 0.53) × 109 | (7.43 ± 4.27) × 108 |
| Chlorophyll a-b binding protein 2 | P12331 | n.d. | (4.84 ± 2.39) × 106 | n.d. | (4.29 ± 4.18) × 106 |
| Chlorophyll a-b binding protein | Q7XV11 | (1.71 ± 1.01) × 106 | (3.83 ± 0.95) × 106 | (1.10 ± 0.44) × 107 | (9.44 ± 4.78) × 106 |
| Ribulose bisphosphate carboxylase large chain | P0C512 | (1.08 ± 0.97) × 107 | n.d. | (1.52 ± 0.29) × 107 | (1.04 ± 0.78) × 107 |
| ATP synthase subunit beta | P12085 | (3.65 ± 2.51) × 106 | n.d. | n.d. | n.d. |
| PLASTID TRANSCRIPTIONALLY ACTIVE 16 | Q0DJF9 | (2.55 ± 1.90) × 107 | (2.05 ± 1.54) × 107 | (0.81 ± 1.17) × 107 | (5.19 ± 7.87) × 107 |
| protein CURVATURE THYLAKOID 1A | Q5Z6P4 | (8.76 ± 1.51) × 107 | (3.11 ± 0.67) × 107 | (2.55 ± 0.04) × 107 | (6.79 ± 4.23) × 107 |
| PLASTID MOVEMENT IMPAIRED1 | Q0IZR7 | n.d. | n.d. | n.d. | (4.16 ± 1.49) × 106 |
| Pyruvate, phosphate dikinase 1 | Q6AVA8 | (3.35 ± 2.78) × 105 | (4.62 ±0.02) × 105 | (1.35 ± 1.24) × 106 | n.d. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inomata, T.; Baslam, M.; Masui, T.; Koshu, T.; Takamatsu, T.; Kaneko, K.; Pozueta-Romero, J.; Mitsui, T. Proteomics Analysis Reveals Non-Controlled Activation of Photosynthesis and Protein Synthesis in a Rice npp1 Mutant under High Temperature and Elevated CO2 Conditions. Int. J. Mol. Sci. 2018, 19, 2655. https://doi.org/10.3390/ijms19092655
Inomata T, Baslam M, Masui T, Koshu T, Takamatsu T, Kaneko K, Pozueta-Romero J, Mitsui T. Proteomics Analysis Reveals Non-Controlled Activation of Photosynthesis and Protein Synthesis in a Rice npp1 Mutant under High Temperature and Elevated CO2 Conditions. International Journal of Molecular Sciences. 2018; 19(9):2655. https://doi.org/10.3390/ijms19092655
Chicago/Turabian StyleInomata, Takuya, Marouane Baslam, Takahiro Masui, Tsutomu Koshu, Takeshi Takamatsu, Kentaro Kaneko, Javier Pozueta-Romero, and Toshiaki Mitsui. 2018. "Proteomics Analysis Reveals Non-Controlled Activation of Photosynthesis and Protein Synthesis in a Rice npp1 Mutant under High Temperature and Elevated CO2 Conditions" International Journal of Molecular Sciences 19, no. 9: 2655. https://doi.org/10.3390/ijms19092655