Tropomyosins: Potential Biomarkers for Urothelial Bladder Cancer
Abstract
:1. Introduction
2. Tropomyosin
2.1. Background and Diversity
2.2. Function
2.3. Splice Form Variants
- TPM1—19 protein-coding isoforms and 6 nonsense-mediated decay (NMD) candidates
- TPM2—8 protein-coding isoforms
- TPM3—12 protein-coding and 2 NMD candidates
- TPM4—14 protein-coding and 5 NMD candidates
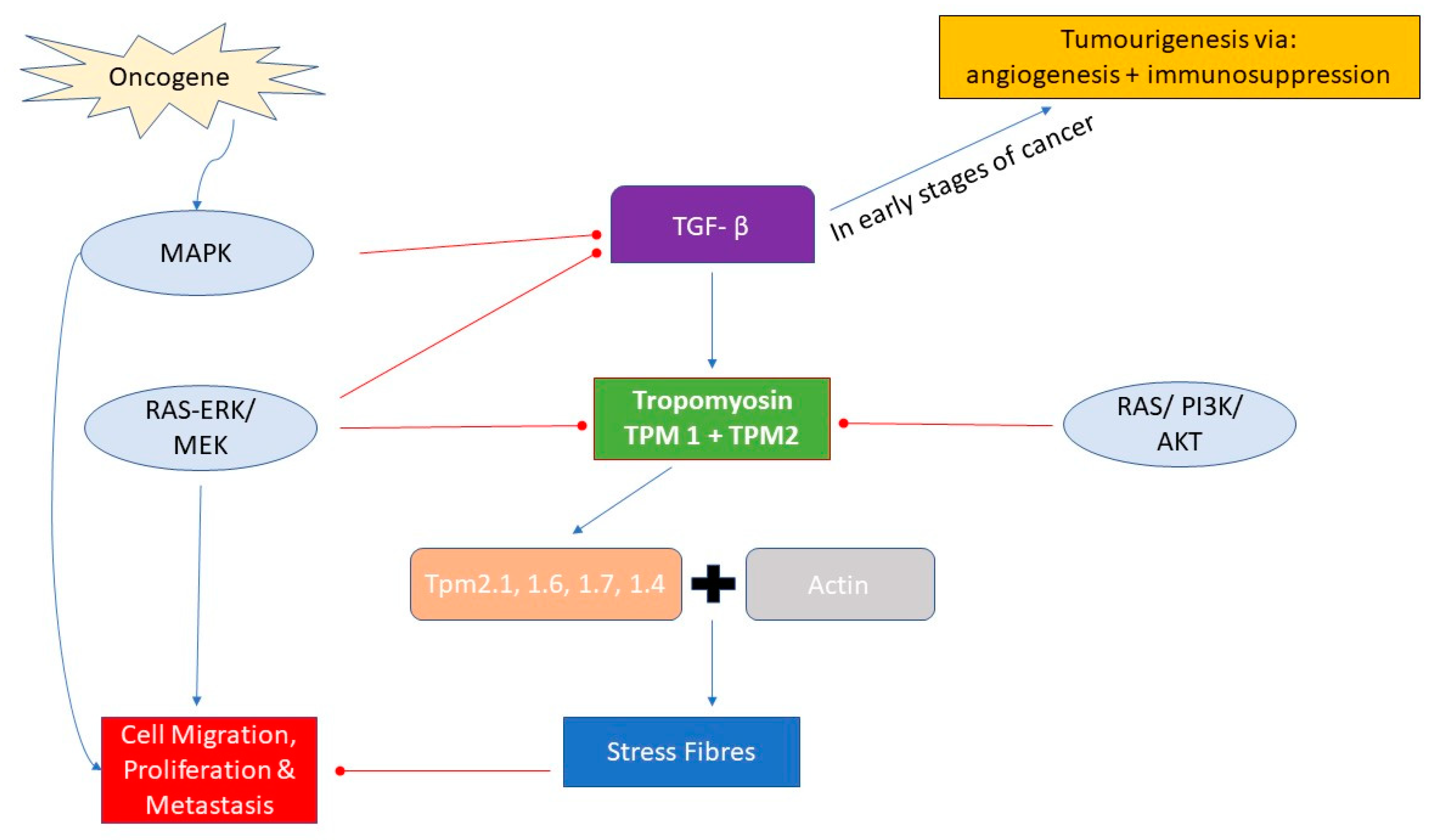
3. TPM Expression and Signalling Pathways in Cancer
4. Tropomyosin and Urothelial Bladder Cancer
4.1. Potential Molecular Mechanisms of Regulation of TPM Isoform Usage
4.2. Isoform Switching Due to Alternative Promoter Usage
- TPM1 has differential presence of RXRA (retinoid X receptor alpha) transcription factor binding-site (TFBS) for its LMW isoforms, as observed in two cell lines: H1-hESC (a human embryonic stem cell line) and HepG2 (a human liver cancer cell line).
- TPM3 and TPM4 have differential presence of the E2F1 TFBS for its LMW isoforms, as observed in the HeLa (cervical cancer) and MCF-7 (a breast cancer cell line) cell lines.
5. TPM as a Marker for Cancer Prognosis
5.1. UBC
5.2. Other Cancers
6. Tropomyosins as Gene Fusion Partners
7. Detection in Urine
8. Tropomyosin Isoforms in Other Cancers
8.1. Malignant Breast Epithelial Cells
8.2. Cholangiocarcinoma
8.3. Prostate Cancer
8.4. Colon Cancer
8.5. Oesophageal Cancer
9. Discussion and Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| UBC | Urothelial Bladder Cancer |
| NMIBC | Non-Muscle Invasive Bladder Cancer |
| MIBC | Muscle Invasive Bladder Cancer |
| TURBT | Transurethral Resection of Bladder Tumour |
| TPM | Tropomyosin (protein) |
| TPM | Tropomyosin gene |
| MAPK | Mitogen Activated Protein Kinase |
References
- Edwards, T.; Dickinson, A.; Natale, S.; Gosling, J.; McGrath, J. A prospective analysis of the diagnostic yield resulting from the attendance of 4020 patients at a protocol-driven haematuria clinic. BJU Int. 2006, 97, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.; Zeegers, M.; van Roekel, E.; Bird, D.; Grant, M.; Dunn, J.; Bathers, S.; Iqbal, G.; Khan, H.; Collins, S.; et al. A comparison of patient and tumour characteristics in two UK bladder cancer cohorts separated by 20 years. BJU Int. 2013, 112, 169–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babjuk, M.; Böhle, A.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.; Hernández, V.; Kaasinen, E.; Palou, J.; Rouprêt, M.; et al. EAU Guidelines on Non–Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur. Urol. 2017, 71, 447–461. [Google Scholar] [CrossRef] [PubMed]
- Mowatt, G.; Zhu, S.; Kilonzo, M.; Boachie, C.; Fraser, C.; Griffiths, T.; N’Dow, J.; Nabi, G.; Cook, J.; Vale, L. Systematic review of the clinical effectiveness and cost-effectiveness of photodynamic diagnosis and urine biomarkers (FISH, ImmunoCyt, NMP22) and cytology for the detection and follow-up of bladder cancer. Health Technol. Assess. 2010, 14. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.; Kirby, R.; O’Brien, T.; Mostafid, H. So Much Cost, Such Little Progress. Eur. Urol. 2014, 66, 263–264. [Google Scholar] [CrossRef] [PubMed]
- Svatek, R.; Hollenbeck, B.; Holmäng, S.; Lee, R.; Kim, S.; Stenzl, A.; Lotan, Y. The Economics of Bladder Cancer: Costs and Considerations of Caring for This Disease. Eur. Urol. 2014, 66, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Campi, R.; Seisen, T.; Roupret, M. Unmet Clinical Needs and Future Perspectives in Non–muscle-invasive Bladder Cancer. Eur. Urol. Focus 2018, 4, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Dräger, B.; Droller, M.; Lokeshwar, V.; Lotan, Y.; Hudson, M.; van Rhijn, B.; Marberger, M.; Fradet, Y.; Hemstreet, G.; Malmstrom, P.; et al. Molecular Markers for Bladder Cancer Screening, Early Diagnosis, and Surveillance: The WHO/ICUD Consensus. Urol. Int. 2015, 94, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Soria, F.; Droller, M.; Lotan, Y.; Gontero, P.; D’Andrea, D.; Gust, K.; Rouprêt, M.; Babjuk, M.; Palou, J.; Shariat, S. An up-to-date catalog of available urinary biomarkers for the surveillance of non-muscle invasive bladder cancer. World J. Urol. 2018, 36, 1981–1995. [Google Scholar] [CrossRef] [PubMed]
- Yossepowitch, O.; Herr, H.; Donat, S. Use of Urinary Biomarkers for Bladder Cancer Surveillance: Patient Perspectives. J. Urol. 2007, 177, 1277–1282. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Lv, Y.; Zhong, Q.; Wang, R.; Jiang, Q. Narrow band imaging diagnosis of bladder cancer: systematic review and meta-analysis. BJU Int. 2012, 110, E680–E687. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K. Tropomyosin: A new asymmetric protein component of the muscle fibril. Biochemistry 1948, 43, 271–279. [Google Scholar] [CrossRef]
- Gunning, P.; O’neill, G.; Hardeman, E. Tropomyosin-Based Regulation of the Actin Cytoskeleton in Time and Space. Phys. Rev. 2008, 88, 1–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; Funke, R.; Gage, D.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Lees, J.; Bach, C.; O’Neill, G. Interior decoration: Tropomyosin in actin dynamics and cell migration. Cell Adh. Migr. 2011, 5, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Geeves, M.A.; Hitchcock-DeGregori, S.E.; Gunning, P.W. A systematic nomenclature for mammalian tropomyosin isoforms. J. Muscle Res. Cell Motil. 2014, 36, 147–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savill, S.; Leitch, H.F.; Harvey, J.N.; Thomas, T.H. Functional structure of the promoter regions for the predominant low molecular weight isoforms of tropomyosin in human kidney cells. J. Cell. Biochem. 2012, 113, 3576–3586. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.J.; Hegmann, T.E.; Lin, J.L. Differential localization of tropomyosin isoforms in cultured nonmuscle cells. J. Cell Biol. 1988, 107, 563–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janco, M.; Bonello, T.T.; Byun, A.; Coster, A.C.; Lebhar, H.; Dedova, I.; Gunning, P.W.; Böcking, T. The impact of tropomyosins on actin filament assembly is isoform specific. BioArchitecture 2016, 6, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Gunning, P.W.; Hardeman, E.C.; Lappalainen, P.; Mulvihill, D.P. Tropomyosin - master regulator of actin filament function in the cytoskeleton. J. Cell Sci. 2015, 128, 2965–2974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhn, T.B.; Bamburg, J.R. Tropomyosin and ADF/Cofilin as Collaborators and Competitors. In Tropomyosin. Advances in Experimental Medicine and Biology; Gunning, P.W., Ed.; Springer: New York, NY, USA, 2008; Volume 644, pp. 232–249. [Google Scholar]
- Ishikawa, R.; Yamashiro, S.; Matsumura, F. Differential Modulation of Actin-severing Activity of Gelsolin by Multiple Isoforms of Cultured Rat Cell Tropomyosin. J. Biol. Chem. 1989, 264, 7490–7497. [Google Scholar] [PubMed]
- Creed, S.J.; Desouza, M.; Bamburg, J.R.; Gunning, P.; Stehn, J. Tropomyosin isoform 3 promotes the formation of filopodia by regulating the recruitment of actin-binding proteins to actin filaments. Exp. Cell Res. 2011, 317, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Schevzov, G.; Vrhovski, B.; Bryce, N.S.; Elmir, S.; Qiu, M.R.; O’Neill, G.M.; Yang, N.; Verrills, N.M.; Kavallaris, M.; Gunning, P.W. Tissue-specific tropomyosin isoform composition. J. Histochem. Cytochem. 2005, 53, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Novy, R.E.; Lin, J.L.; Lin, C.S.; Lin, J.J. Human fibroblast tropomyosin isoforms: Characterization of cDNA clones and analysis of tropomyosin isoform expression in human tissues and in normal and transformed cells. Cell Motil. Cytoskeleton 1993, 25, 267–281. [Google Scholar] [CrossRef] [PubMed]
- Bakin, A.; Safina, A.; Rinehart, C.; Daroqui, C.; Darbary, H.; Helfman, D. A Critical Role of Tropomyosins in TGF-β Regulation of the Actin Cytoskeleton and Cell Motility in Epithelial Cells. Mol. Biol. Cell 2004, 15, 4682–4694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabregat, I.; Fernando, J.; Mainez, J.; Sancho, P. TGF-beta Signaling in Cancer Treatment. Curr. Pharm. Des. 2014, 20, 2934–2947. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Wakefield, L. The two faces of transforming growth factor in carcinogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8621–8623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedegaard, J.; Lamy, P.; Nordentoft, I.; Algaba, F.; Høyer, S.; Ulhøi, B.P.; Vang, S.; Reinert, T.; Hermann, G.G.; Mogensen, K.; et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 2016, 30, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Bryan, R.; Hussain, S.; James, N.; Jankowski, J.; Wallace, D. Molecular pathways in bladder cancer: Part 1. BJU Int. 2005, 95, 485–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bryan, R.; Hussain, S.; James, N.; Jankowski, J.; Wallace, D. Molecular pathways in bladder cancer: Part 2. BJU Int. 2005, 95, 491–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlak, G.; McGarvey, T.W.; Nguyen, T.B.; Tomaszewski, J.E.; Puthiyaveettil, R.; Malkowicz, S.B.; Helfman, D.M. Alterations in tropomyosin isoform expression in human transitional cell carcinoma of the urinary bladder. Int. J. Cancer 2004, 110, 368–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Zhao, X.; Zhou, J.; Cheng, X.; Ye, Z.; Ji, Z. Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol. Ther. 2018, 19, 1039–1056. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Feng, C.; Xu, Y. Cyclin-dependent kinase-associated protein Cks2 is associated with bladder cancer progression. J. Int. Med. Res. 2011, 39, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Kee, A.J.; Yang, L.; Lucas, C.A.; Greenberg, M.J.; Martel, N.; Leong, G.M.; Hughes, W.E.; Cooney, G.J.; James, D.E.; Ostap, E.M.; et al. An actin filament population defined by the tropomyosin Tpm3.1 regulates glucose uptake. Traffic 2015, 16, 691–711. [Google Scholar] [CrossRef] [PubMed]
- Bryce, N.S.; Schevzov, G.; Ferguson, V.; Percival, J.M.; Lin, J.J.; Matsumura, F.; Bamburg, J.R.; Jeffrey, P.L.; Hardeman, E.C.; Gunning, P.; et al. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 2003, 14, 1002–1016. [Google Scholar] [CrossRef] [PubMed]
- Brayford, S.; Bryce, N.; Schevzov, G.; Haynes, E.; Bear, J.; Hardeman, E.; Gunning, P. Tropomyosin Promotes Lamellipodial Persistence by Collaborating with Arp2/3 at the Leading Edge. Curr. Biol. 2016, 26, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Lerner, S.P.; McConkey, D.J.; Hoadley, K.A.; Chan, K.S.; Kim, W.Y.; Radvanyi, F.; Höglund, M.; Real, F.X. Bladder Cancer Molecular Taxonomy: Summary from a consensus meeting. Bladder Cancer 2016, 2, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Leem, S.H.; Lee, S.Y.; Kim, S.C.; Park, E.S.; Kim, S.B.; Kim, S.K.; Kim, Y.J.; Kim, W.J.; Chu, I.S. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. J. Clin. Oncol. 2010, 28, 2660–2667. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Shi, Y.; Han, D.; Luan, W.; Qian, J.; Zhang, J.; Wang, Y.; You, Y. TPM3, a strong prognosis predictor, is involved in malignant progression through MMP family members and EMT-like activators in gliomas. Tumour Biol. 2014, 35, 9053–9059. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Yim, S.; Xu, H.; Jung, S.; Shin, S.; Hu, H.; Jung, C.; Choi, J.; Chung, Y. Tropomyosin3 overexpression and a potential link to epithelial-mesenchymal transition in human hepatocellular carcinoma. BMC Cancer 2010, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- WANG, J.; GUAN, J.; LU, Z.; JIN, J.; CAI, Y.; WANG, C.; WANG, F. Clinical and tumor significance of tropomyosin-1 expression levels in renal cell carcinoma. Oncol. Rep. 2015, 33, 1326–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gong, M.; Yuan, H.; Park, H.; Frierson, H.; Li, H. Chimeric Transcript Generated by cis-Splicing of Adjacent Genes Regulates Prostate Cancer Cell Proliferation. Cancer Discov. 2012, 2, 598–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariño-Enríquez, A.; Dal Cin, P. ALK as a paradigm of oncogenic promiscuity: different mechanisms of activation and different fusion partners drive tumors of different lineages. Cancer Genet. 2013, 206, 357–373. [Google Scholar] [CrossRef] [PubMed]
- de Alava, E.; Kawai, A.; Healey, J.; Fligman, I.; Meyers, P.; Huvos, A.; Gerald, W.; Jhanwar, S.; Argani, P.; Antonescu, C.; et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin. Oncol. 1998, 16, 1248–1255. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, B.; Perez-Atayde, A.; Hibbard, M.; Rubin, B.; Dal Cin, P.; Pinkus, J.; Pinkus, G.; Xiao, S.; Yi, E.; Fletcher, C.; et al. TPM3-ALK and TPM4-ALK Oncogenes in Inflammatory Myofibroblastic Tumors. Am. J Pathol. 2000, 157, 377–384. [Google Scholar] [CrossRef]
- Stransky, N.; Cerami, E.; Schalm, S.; Kim, J.; Lengauer, C. The landscape of kinase fusions in cancer. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.; Bamford, S.; Jubb, H.; Sondka, Z.; Beare, D.; Bindal, N.; Boutselakis, H.; Cole, C.; Creatore, C.; Dawson, E.; et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2018, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Soda, M.; Togashi, Y.; Suzuki, R.; Sakata, S.; Hatano, S.; Asaka, R.; Hamanaka, W.; Ninomiya, H.; Uehara, H.; et al. RET, ROS1 and ALK fusions in lung cancer. Nat. Med. 2012, 18, 378–381. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, T.; He, J.; Yelensky, R.; Esteve-Puig, R.; Botton, T.; Yeh, I.; Lipson, D.; Otto, G.; Brennan, K.; Murali, R.; et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Habib, H.; Ali, E.; Kotb, Y. Evaluation of urinary miRNA-96 as a potential biomarker for bladder cancer diagnosis. Med. Oncol. 2014, 32. [Google Scholar] [CrossRef] [PubMed]
- Sin, M.; Mach, K.; Sinha, R.; Wu, F.; Trivedi, D.; Altobelli, E.; Jensen, K.; Sahoo, D.; Lu, Y.; Liao, J. Deep Sequencing of Urinary RNAs for Bladder Cancer Molecular Diagnostics. Clin. Cancer Res. 2017, 23, 3700–3710. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, M.; Yang, Y.; Guo, Z.; Sun, Y.; Shao, C.; Li, M.; Sun, W.; Gao, Y. A comprehensive analysis and annotation of human normal urinary proteome. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.; Borràs, F.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.; Yalamanchili, S.; Lachant, J.; Abbott, L.; Benz, P.; Mitschow, C.; Dube, D.; Poiesz, B. Expression of Tropomyosin 1 Gene Isoforms in Human Breast Cancer Cell Lines. Int. J. Breast Cancer 2015, 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dube, S.; Thomas, A.; Abbott, L.; Benz, P.; Mitschow, C.; Dube, D.; Poiesz, B. Expression of tropomyosin 2 gene isoforms in human breast cancer cell lines. Oncol. Rep. 2016, 35, 3143–3150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Wang, X.; Zheng, W.; Li, K.; Liu, H.; Sun, Y. Genetic and epigenetic alterations are involved in the regulation of TPM1 in cholangiocarcinoma. Int. J. Oncol. 2012, 42, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Thorsen, K.; Sørensen, K.; Brems-Eskildsen, A.; Modin, C.; Gaustadnes, M.; Hein, A.; Kruhøffer, M.; Laurberg, S.; Borre, M.; Wang, K.; et al. Alternative Splicing in Colon, Bladder, and Prostate Cancer Identified by Exon Array Analysis. Mol. Cell. Proteom. 2008, 7, 1214–1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Geng, X.; Bhattacharya, S.; Yu, J.; Reiter, R.; Sastri, B.; Glazier, K.; Mirza, Z.; Wang, K.; Amenta, P.; et al. Isolation and sequencing of a novel tropomyosin isoform preferentially associated with colon cancer. Gastroenterology 2002, 123, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Zare, M.; Jazii, F.; Soheili, Z.; Moghanibashi, M. Downregulation of tropomyosin-1 in squamous cell carcinoma of esophagus, the role of Ras signaling and methylation. Mol. Carcinog. 2011, 51, 796–806. [Google Scholar] [CrossRef] [PubMed]

| Tropomyosin Gene | TPM Isoforms Common (Old) Name | TPM Nomenclature | Size (HMW/LMW) | Molecular Weight (kDa) |
|---|---|---|---|---|
| TPM1 | Tm2 Tm3 Tm5a Tm5b Tm6 | Tpm 1.6 Tpm 1.7 Tpm 1.8 Tpm 1.9 Tpm 1.4 | HMW HMW LMW LMW HMW | 36 34 30 30 40 |
| TPM2 | Tm1 | Tpm 2.1 | HMW | 38 |
| TPM3 | Tm5NM1 | Tpm 3.1 | LMW | 30 |
| TPM4 | Tm4.1 Tm4 | Tpm 4.1 Tpm 4.2 | HMW LMW | 30 |
| Bladder Cancer Stage | Hallmark | Process | TPM Involvement |
|---|---|---|---|
| Normal | Self-sufficiency in growth signals | Normal expression levels of TPM genes | Tpm 1.6 and Tpm 1.7—formation of stress fibres, cell stability, reduced motility |
| Suppression | Insensitivity to growth inhibitory signals | Cell-cycle arrest in G0/G1 if proliferating too much TPM1 + MEG3 causes apoptosis by upregulation of Bcl2 and caspase3 | TPM1—acts as tumour suppressor |
| Initiation | Evasion of programmed cell death | Decreased expression of TPM1 and TPM2 TPM1 is downregulated due to increased micro-RNA 96 (suppression lost) | Loss of TPM1—EMT Tpm 2.1—responsible for anoikis, maintains cell–cell adhesion |
| Non-invasive | Limitless replicative potential | Mutagenic MAPK signalling activated | Tpm3.1—cellular growth, proliferation and motility |
| Invasive | Sustained angiogenesis | Invasion into extracellular matrix (ECM) Angiogenesis stimulated by concurrent pathways | LMW TPMs—formation of lamellipodia, increased cell motility and morphogenesis |
| Metastasis | Tissue invasion and death | Breakthrough ECM with distant spread | LMW isoforms of TPM3 and TPM4—Cancer cell survival, focal adhesion, MEK/ERK-mediated proliferation |
| Evading immune destruction | Ectopic expression of TPM1 and TPM2 Continued uninhibited anchorage-independent growth |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Humayun-Zakaria, N.; Arnold, R.; Goel, A.; Ward, D.; Savill, S.; Bryan, R.T. Tropomyosins: Potential Biomarkers for Urothelial Bladder Cancer. Int. J. Mol. Sci. 2019, 20, 1102. https://doi.org/10.3390/ijms20051102
Humayun-Zakaria N, Arnold R, Goel A, Ward D, Savill S, Bryan RT. Tropomyosins: Potential Biomarkers for Urothelial Bladder Cancer. International Journal of Molecular Sciences. 2019; 20(5):1102. https://doi.org/10.3390/ijms20051102
Chicago/Turabian StyleHumayun-Zakaria, Nada, Roland Arnold, Anshita Goel, Douglas Ward, Stuart Savill, and Richard T. Bryan. 2019. "Tropomyosins: Potential Biomarkers for Urothelial Bladder Cancer" International Journal of Molecular Sciences 20, no. 5: 1102. https://doi.org/10.3390/ijms20051102





