Sonic Hedgehog-Gli1 Signaling and Cellular Retinoic Acid Binding Protein 1 Gene Regulation in Motor Neuron Differentiation and Diseases
Abstract
:1. Introduction
2. Results
2.1. Crabp1 is Highly Expressed in Spinal Motor Neurons
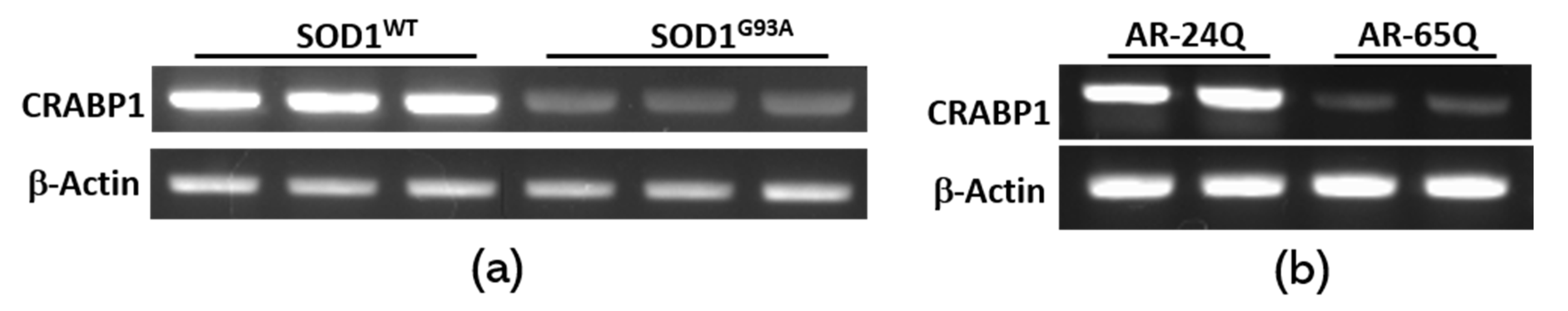
2.2. Crabp1 Expression is Down-Regulated in Diseased Motor Neurons
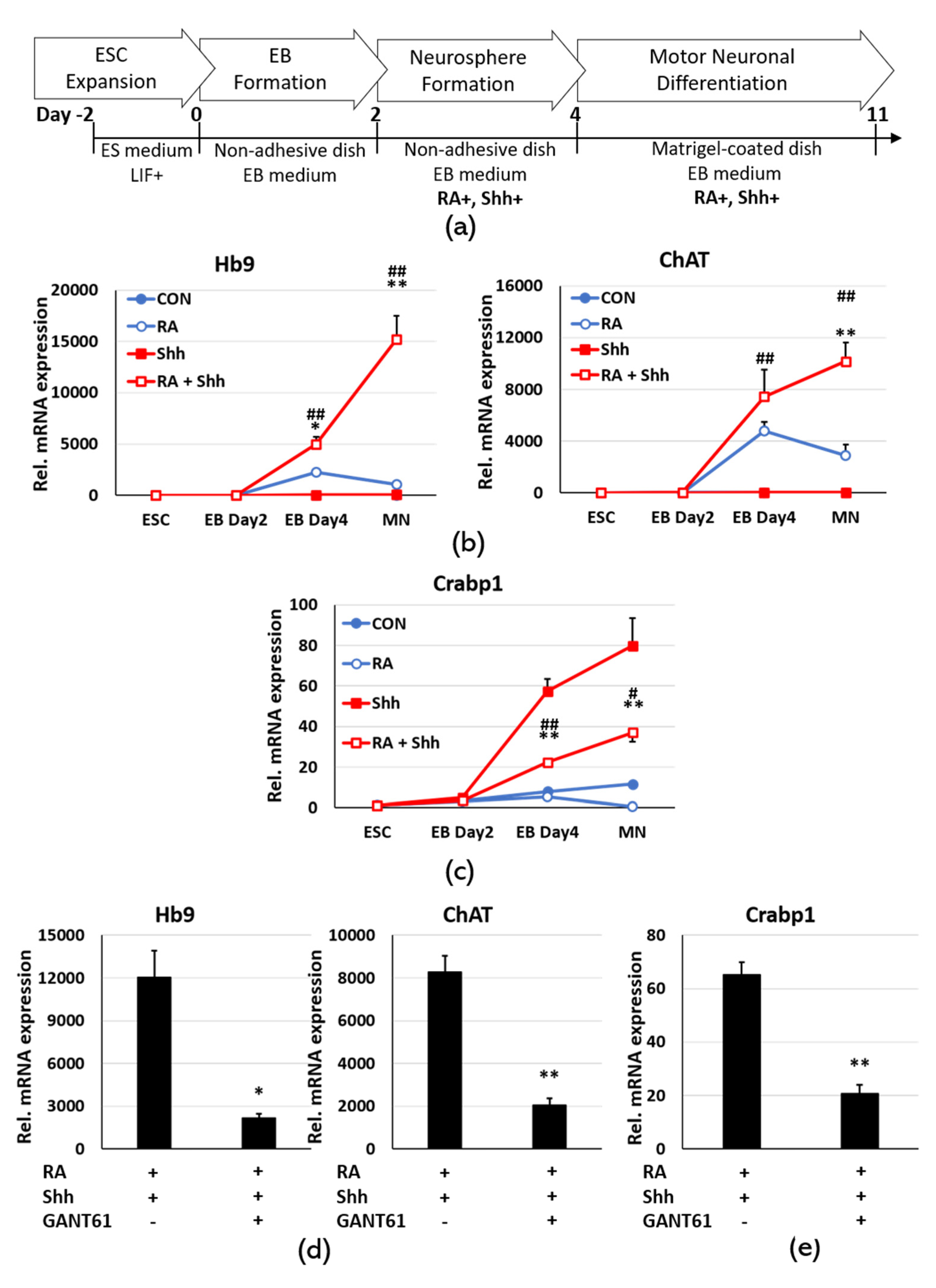
2.3. Sonic Hedgehog Signaling Up-Regulates Crabp1 in Motor Neuron Differentiation
2.4. Gli1 Directly Binds to Its Chromatin Target on Crabp1, Inducing Juxtaposition with the Minimal Promoter to Up-regulate Crabp1 Expression
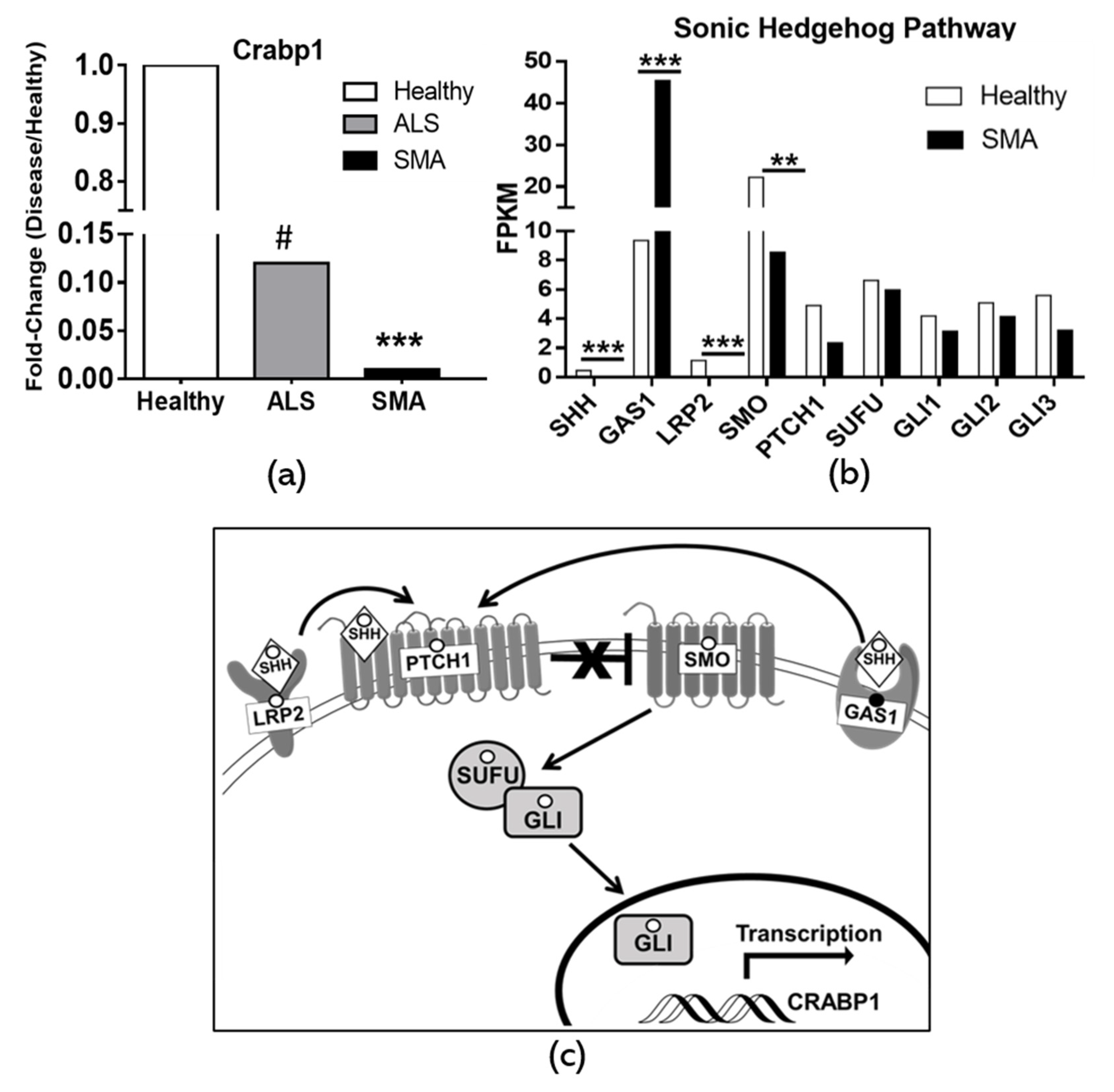
2.5. Dysregulation of CRABP1 and Shh Signaling Components in Human Motor Neuron Diseases
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Western Blotting
4.3. Immunohistochemistry
4.4. Cell Culture and Disease Models
4.5. ESCs Culture and Differentiation into Motor Neurons
4.6. Quantitative Real-time PCR (qPCR)
4.7. Chromatin Immunoprecipitation (ChIP) Assay
4.8. Data Mining of crabp1 and Shh in Human Motor Neuron Disease
4.9. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic lateral sclerosis |
| AR | Androgen receptor |
| CKO | Crabp1 knockout |
| CNS | Central nervous system |
| Crabp1 | Cellular retinoic acid-binding protein 1 |
| ESC | Embryonic stem cell |
| FKPM | Fragments per kilobase of transcript per million |
| Gli | Glioma-associated oncogene homolog |
| HRE | Hormone response element |
| iPSC | Induced pluripotent stem cell |
| PIC | Preinitiation complex |
| RA | Retinoic acid |
| SBMA | Spinal and bulbar muscular atrophy |
| Shh | Sonic hedgehog |
| SMA | Spinal muscular atrophy |
| SMO | Smoothened homolog |
| TIS | Transcription initiation site |
| WT | Wild type |
References
- Wei, L.N.; Chang, L.; Hu, X. Studies of the type I cellular retinoic acid-binding protein mutants and their biological activities. Mol. Cell. Biochem. 1999, 200, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Fiorella, P.D.; Napoli, J.L. Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J. Biol. Chem. 1994, 269, 10538–10544. [Google Scholar] [PubMed]
- Napoli, J.L. Retinoic acid: Its biosynthesis and metabolism. Prog. Nucleic Acid Res. Mol. Biol. 1999, 63, 139–188. [Google Scholar]
- Perez-Castro, A.V.; Toth-Rogler, L.E.; Wei, L.N.; Nguyen-Huu, M.C. Spatial and temporal pattern of expression of the cellular retinoic acid-binding protein and the cellular retinol-binding protein during mouse embryogenesis. Proc. Natl. Acad. Sci. USA 1989, 86, 8813–8817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.N.; Chen, G.J.; Chu, Y.S.; Tsao, J.L.; Nguyen-Huu, M.C. A 3 kb sequence from the mouse cellular retinoic-acid-binding protein gene upstream region mediates spatial and temporal LacZ expression in transgenic mouse embryos. Development 1991, 112, 847–854. [Google Scholar] [PubMed]
- Wei, L.N.; Tsao, J.L.; Chu, Y.S.; Jeannotte, L.; Nguyen-Huu, M.C. Molecular cloning and transcriptional mapping of the mouse cellular retinoic acid-binding protein gene. DNA Cell Biol. 1990, 9, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Huang, W.H.; Persaud, S.D.; Wei, L.N. RIP140 in thyroid hormone-repression and chromatin remodeling of Crabp1 gene during adipocyte differentiation. Nucleic Acids Res. 2009, 37, 7085–7094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.N.; Lee, C.H. Demethylation in the 5′-flanking region of mouse cellular retinoic acid binding protein-I gene is associated with its high level of expression in mouse embryos and facilitates its induction by retinoic acid in P19 embryonal carcinoma cells. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1994, 201, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Li, G.; Lin, Y.P.; Barrero, M.J.; Ge, K.; Roeder, R.G.; Wei, L.N. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell 2005, 19, 643–653. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Wei, L.N. Characterization of a negative response DNA element in the upstream region of the cellular retinoic acid-binding protein-I gene of the mouse. J. Biol. Chem. 1997, 272, 10144–10150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.N.; Chang, L. Promoter and upstream regulatory activities of the mouse cellular retinoic acid-binding protein-I gene. J. Biol. Chem. 1996, 271, 5073–5078. [Google Scholar] [PubMed] [Green Version]
- Wei, L.N.; Lee, C.H.; Chang, L. Retinoic acid induction of mouse cellular retinoic acid-binding protein-I gene expression is enhanced by sphinganine. Mol. Cell. Endocrinol. 1995, 111, 207–211. [Google Scholar] [CrossRef]
- Lin, Y.W.; Park, S.W.; Lin, Y.L.; Burton, F.H.; Wei, L.N. Cellular retinoic acid binding protein 1 protects mice from high-fat diet-induced obesity by decreasing adipocyte hypertrophy. Int J. Obes. (L.) 2020, 44, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Nhieu, J.; Lin, Y.W.; Wei, L.N. All-trans retinoic acid attenuates isoproterenol-induced cardiac dysfunction through Crabp1 to dampen CaMKII activation. Eur. J. Pharmacol. 2019, 858, 172485. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Persaud, S.D.; Nhieu, J.; Wei, L.N. Cellular Retinoic Acid-Binding Protein 1 Modulates Stem Cell Proliferation to Affect Learning and Memory in Male Mice. Endocrinology 2017, 158, 3004–3014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, M.; Harris, A.W.; Kingham, B.F.; Lumpkin, C.J.; Opdenaker, L.M.; McCahan, S.M.; Wang, W.; Butchbach, M.E. Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PLoS ONE 2014, 9, e106818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.M.; Yamamoto, M.; Kobayashi, Y.; Yoshihara, T.; Liang, Y.; Terao, S.; Takeuchi, H.; Ishigaki, S.; Katsuno, M.; Adachi, H.; et al. Gene expression profile of spinal motor neurons in sporadic amyotrophic lateral sclerosis. Ann. Neurol. 2005, 57, 236–251. [Google Scholar] [CrossRef] [PubMed]
- Swindell, W.R.; Bojanowski, K.; Kindy, M.S.; Chau, R.M.W.; Ko, D. GM604 regulates developmental neurogenesis pathways and the expression of genes associated with amyotrophic lateral sclerosis. Transl. Neurodegener. 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.C.; Wei, L.N. Expression of cellular retinoic acid-binding protein I is specific to neurons in adult transgenic mouse brain. Brain Res. Gene Expr. Patterns 2001, 1, 67–72. [Google Scholar] [CrossRef]
- Chen, S.D.; Yang, J.L.; Hwang, W.C.; Yang, D.I. Emerging Roles of Sonic Hedgehog in Adult Neurological Diseases: Neurogenesis and Beyond. Int. J. Mol. Sci. 2018, 19, 2423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, S.J.; McKeown, S.R.; Rashid, S. Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene 2016, 577, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Grunseich, C.; Zukosky, K.; Kats, I.R.; Ghosh, L.; Harmison, G.G.; Bott, L.C.; Rinaldi, C.; Chen, K.L.; Chen, G.; Boehm, M.; et al. Stem cell-derived motor neurons from spinal and bulbar muscular atrophy patients. Neurobiol. Dis. 2014, 70, 12–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liyang, G.; Abdullah, S.; Rosli, R.; Nordin, N. Neural Commitment of Embryonic Stem Cells through the Formation of Embryoid Bodies (EBs). Malays. J. Med. Sci. 2014, 21, 8–16. [Google Scholar] [PubMed]
- Guan, K.; Chang, H.; Rolletschek, A.; Wobus, A.M. Embryonic stem cell-derived neurogenesis. Retinoic acid induction and lineage selection of neuronal cells. Cell Tissue Res. 2001, 305, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Wichterle, H.; Lieberam, I.; Porter, J.A.; Jessell, T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell 2002, 110, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Kasaai, B.; Gaumond, M.H.; Moffatt, P. Regulation of the bone-restricted IFITM-like (Bril) gene transcription by Sp and Gli family members and CpG methylation. J. Biol. Chem. 2013, 288, 13278–13294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Gao, L.; Jia, H.; Bai, Y.; Wang, W. The Expression of Shh, Ptch1, and Gli1 in the Developing Caudal Spinal Cord of Fetal Rats with Anorectal Malformations. J. Surg. Res. 2019, 233, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, F.; Nizzardo, M.; Vashisht, S.; Molteni, E.; Melzi, V.; Taiana, M.; Salani, S.; Santonicola, P.; Di Schiavi, E.; Bucchia, M.; et al. Key role of SMN/SYNCRIP and RNA-Motif 7 in spinal muscular atrophy: RNA-Seq and motif analysis of human motor neurons. Brain 2019, 142, 276–294. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Zhang, W.; Krauss, R.S. Hedgehog signaling: Cooking with Gas1. Sci. Signal. 2007, 2007, 50. [Google Scholar] [CrossRef] [PubMed]
- Gorry, P.; Lufkin, T.; Dierich, A.; Rochette-Egly, C.; Decimo, D.; Dolle, P.; Mark, M.; Durand, B.; Chambon, P. The cellular retinoic acid binding protein I is dispensable. Proc. Natl. Acad. Sci. USA 1994, 91, 9032–9036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bruijn, D.R.; Oerlemans, F.; Hendriks, W.; Baats, E.; Ploemacher, R.; Wieringa, B.; Geurts van Kessel, A. Normal development, growth and reproduction in cellular retinoic acid binding protein-I (CRABPI) null mutant mice. Differ. Res. Biol. Divers. 1994, 58, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Nhieu, J.; Persaud, S.D.; Miller, M.C.; Xia, Y.; Lin, Y.W.; Lin, Y.L.; Kagechika, H.; Mayo, K.H.; Wei, L.N. A new regulatory mechanism for Raf kinase activation, retinoic acid-bound Crabp1. Sci. Rep. 2019, 9, 10929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Persaud, S.D.; Park, S.W.; Ishigami-Yuasa, M.; Koyano-Nakagawa, N.; Kagechika, H.; Wei, L.N. All trans-retinoic acid analogs promote cancer cell apoptosis through non-genomic Crabp1 mediating ERK1/2 phosphorylation. Sci. Rep. 2016, 6, 22396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janesick, A.; Wu, S.C.; Blumberg, B. Retinoic acid signaling and neuronal differentiation. Cell. Mol. Life Sci. 2015, 72, 1559–1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maden, M. Retinoids and spinal cord development. J. Neurobiol. 2006, 66, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, P.; Lindsey, B.W.; Leopold, C.; Rafuse, V.F. Easy and rapid differentiation of embryonic stem cells into functional motoneurons using sonic hedgehog-producing cells. Stem Cells 2007, 25, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Belgacem, Y.H.; Hamilton, A.M.; Shim, S.; Spencer, K.A.; Borodinsky, L.N. The Many Hats of Sonic Hedgehog Signaling in Nervous System Development and Disease. J. Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.J.; Petralia, R.S.; Mattson, M.P. Sonic Hedgehog Signaling and Hippocampal Neuroplasticity. Trends Neurosci. 2016, 39, 840–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, K.; Kaspar, B.K.; Gage, F.H.; Schaffer, D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003, 6, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Ericson, J.; Morton, S.; Kawakami, A.; Roelink, H.; Jessell, T.M. Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 1996, 87, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.L.; Wang, S. Prenatal lipopolysaccharide exposure increases depression-like behaviors and reduces hippocampal neurogenesis in adult rats. Behav. Brain Res. 2014, 259, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-L.; Lin, Y.-W.; Nhieu, J.; Zhang, X.; Wei, L.-N. Sonic Hedgehog-Gli1 Signaling and Cellular Retinoic Acid Binding Protein 1 Gene Regulation in Motor Neuron Differentiation and Diseases. Int. J. Mol. Sci. 2020, 21, 4125. https://doi.org/10.3390/ijms21114125
Lin Y-L, Lin Y-W, Nhieu J, Zhang X, Wei L-N. Sonic Hedgehog-Gli1 Signaling and Cellular Retinoic Acid Binding Protein 1 Gene Regulation in Motor Neuron Differentiation and Diseases. International Journal of Molecular Sciences. 2020; 21(11):4125. https://doi.org/10.3390/ijms21114125
Chicago/Turabian StyleLin, Yu-Lung, Yi-Wei Lin, Jennifer Nhieu, Xiaoyin Zhang, and Li-Na Wei. 2020. "Sonic Hedgehog-Gli1 Signaling and Cellular Retinoic Acid Binding Protein 1 Gene Regulation in Motor Neuron Differentiation and Diseases" International Journal of Molecular Sciences 21, no. 11: 4125. https://doi.org/10.3390/ijms21114125




