Endocannabinoid System Regulation in Female Rats with Recurrent Episodes of Binge Eating
Abstract
:1. Introduction
2. Results
2.1. BE Behavior in Rats Exposed to Food Restrictions Plus Stress
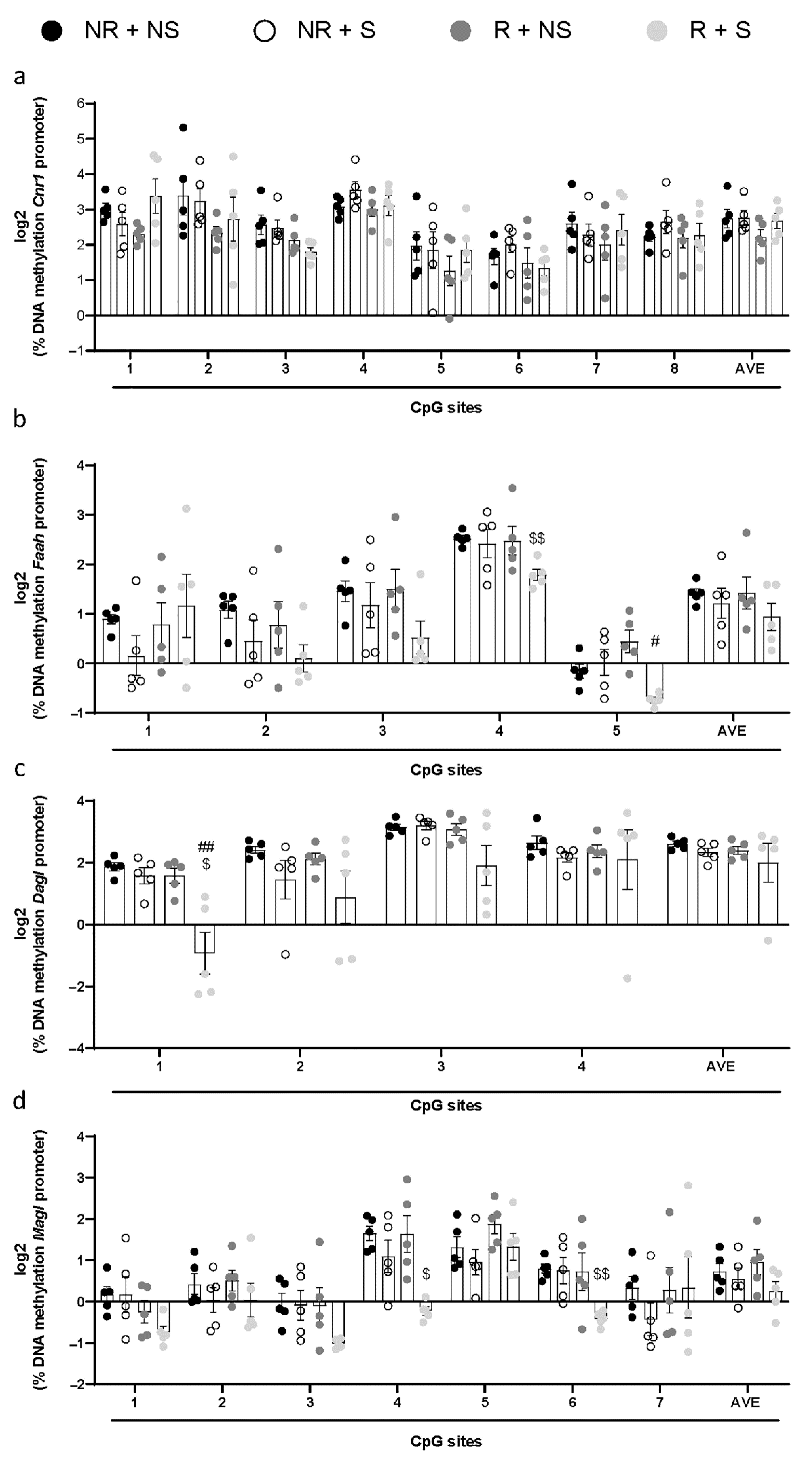
2.2. Regulation of ECS in Rats Exposed to Food Restrictions and Stress Procedure
2.3. Endogenous Content of eCBs
3. Discussion
4. Materials and Methods
4.1. Subjects and Diet Composition
4.2. Tissue Collection, Nucleic Acids and Chromatin Extraction
4.3. Analysis of Gene Expression
4.4. DNA Methylation
4.5. Chromatin Immunoprecipitation
4.6. UHPLC-MS/MS Analysis of eCBs Levels
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kessler, R.M.; Hutson, P.H.; Herman, B.K.; Potenza, M.N. The Neurobiological Basis of Binge-Eating Disorder. Neurosci. Biobehav. Rev. 2016, 63, 223–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- American Psychiatric Association DSM-5 Diagnostic Classification. In Diagnostic and Statistical Manual of Mental Disorders; American Psychiatric Association: Washington, DC, USA, 2013.
- Rodgers, R.F.; Lombardo, C.; Cerolini, S.; Franko, D.L.; Omori, M.; Fuller-Tyszkiewicz, M.; Linardon, J.; Courtet, P.; Guillaume, S. The Impact of the COVID-19 Pandemic on Eating Disorder Risk and Symptoms. Int. J. Eat. Disord. 2020, 53, 1166–1170. [Google Scholar] [CrossRef] [PubMed]
- Cardi, V.; Leppanen, J.; Treasure, J. The Effects of Negative and Positive Mood Induction on Eating Behaviour: A Meta-Analysis of Laboratory Studies in the Healthy Population and Eating and Weight Disorders. Neurosci. Biobehav. Rev. 2015, 57, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Weissman, R.S.; Bauer, S.; Thomas, J.J. Access to Evidence-Based Care for Eating Disorders during the COVID-19 Crisis. Int. J. Eat. Disord. 2020, 53, 369–376. [Google Scholar] [CrossRef]
- Galmiche, M.; Déchelotte, P.; Lambert, G.; Tavolacci, M.P. Prevalence of Eating Disorders over the 2000–2018 Period: A Systematic Literature Review. Am. J. Clin. Nutr. 2019, 109, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Davis, C. The Epidemiology and Genetics of Binge Eating Disorder (BED). CNS Spectr. 2015, 20, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Pearl, R.L.; White, M.A.; Grilo, C.M. Weight Bias Internalization, Depression, and Self-Reported Health among Overweight Binge Eating Disorder Patients. Obesity (Silver Spring) 2014, 22, E142–E148. [Google Scholar] [CrossRef] [Green Version]
- Tammen, S.A.; Friso, S.; Choi, S.-W. Epigenetics: The Link between Nature and Nurture. Mol. Asp. Med. 2013, 34, 753–764. [Google Scholar] [CrossRef] [Green Version]
- Bulik, C.; Yilmaz, Z.; HArdaway, A. Genetics and Epigenetics of Eating Disorders. Adv. Genom. Genet. 2015, 5, 131. [Google Scholar] [CrossRef] [Green Version]
- Micioni Di Bonaventura, M.V.; Pucci, M.; Giusepponi, M.E.; Romano, A.; Lambertucci, C.; Volpini, R.; Micioni Di Bonaventura, E.; Gaetani, S.; Maccarrone, M.; D’Addario, C.; et al. Regulation of Adenosine A2A Receptor Gene Expression in a Model of Binge Eating in the Amygdaloid Complex of Female Rats. J. Psychopharmacol. 2019, 33, 1550–1561. [Google Scholar] [CrossRef]
- Pucci, M.; Micioni Di Bonaventura, M.V.; Giusepponi, M.E.; Romano, A.; Filaferro, M.; Maccarrone, M.; Ciccocioppo, R.; Cifani, C.; D’Addario, C. Epigenetic Regulation of Nociceptin/Orphanin FQ and Corticotropin-Releasing Factor System Genes in Frustration Stress-Induced Binge-like Palatable Food Consumption. Addict. Biol. 2016, 21, 1168–1185. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.; Micioni Di Bonaventura, M.V.; Zaplatic, E.; Bellia, F.; Maccarrone, M.; Cifani, C.; D’Addario, C. Transcriptional Regulation of the Endocannabinoid System in a Rat Model of Binge-Eating Behavior Reveals a Selective Modulation of the Hypothalamic Fatty Acid Amide Hydrolase Gene. Int. J. Eat. Disord. 2019, 52, 51–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, I.C.; Mill, J.; Uher, R.; Schmidt, U. Eating Disorders, Gene-Environment Interactions and Epigenetics. Neurosci. Biobehav. Rev. 2011, 35, 784–793. [Google Scholar] [CrossRef] [PubMed]
- D’Addario, C.; Zaplatic, E.; Giunti, E.; Pucci, M.; Micioni Di Bonaventura, M.V.; Scherma, M.; Dainese, E.; Maccarrone, M.; Nilsson, I.A.; Cifani, C.; et al. Epigenetic Regulation of the Cannabinoid Receptor CB1 in an Activity-Based Rat Model of Anorexia Nervosa. Int. J. Eat. Disord. 2020, 53, 702–716. [Google Scholar] [CrossRef]
- Jager, G.; Witkamp, R.F. The Endocannabinoid System and Appetite: Relevance for Food Reward. Nutr. Res. Rev. 2014, 27, 172–185. [Google Scholar] [CrossRef] [Green Version]
- D’Addario, C.; Micioni Di Bonaventura, M.V.; Pucci, M.; Romano, A.; Gaetani, S.; Ciccocioppo, R.; Cifani, C.; Maccarrone, M. Endocannabinoid Signaling and Food Addiction. Neurosci. Biobehav. Rev. 2014, 47, 203–224. [Google Scholar] [CrossRef]
- Meccariello, R.; Santoro, A.; D’Angelo, S.; Morrone, R.; Fasano, S.; Viggiano, A.; Pierantoni, R. The Epigenetics of the Endocannabinoid System. Int. J. Mol. Sci. 2020, 21, 1113. [Google Scholar] [CrossRef]
- Maccarrone, M. Missing Pieces to the Endocannabinoid Puzzle. Trends Mol. Med. 2020, 26, 263–272. [Google Scholar] [CrossRef]
- Pucci, M.; Zaplatic, E.; Di Bonaventura, M.V.M.; Di Bonaventura, E.M.; De Cristofaro, P.; Maccarrone, M.; Cifani, C.; D’Addario, C. On the Role of Central Type-1 Cannabinoid Receptor Gene Regulation in Food Intake and Eating Behaviors. Int. J. Mol. Sci. 2021, 22, 398. [Google Scholar] [CrossRef]
- Capasso, A.; Milano, W.; Cauli, O. Changes in the Peripheral Endocannabinoid System as a Risk Factor for the Development of Eating Disorders. Endocr. Metab. Immune Disord. Drug Targets 2018, 18, 325–332. [Google Scholar] [CrossRef]
- Monteleone, A.M.; Piscitelli, F.; Dalle Grave, R.; El Ghoch, M.; Di Marzo, V.; Maj, M.; Monteleone, P. Peripheral Endocannabinoid Responses to Hedonic Eating in Binge-Eating Disorder. Nutrients 2017, 9, 1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagin, N.L.; Aliasgari, F.; Alizadeh, M.; Aliasgharzadeh, S.; Mahdavi, R. Comparison of Endocannabinoids Levels, FAAH Gene Polymorphisms, and Appetite Regulatory Substances in Women with and without Binge Eating Disorder: A Cross-Sectional Study. Nutr. Res. 2020, 83, 86–93. [Google Scholar] [CrossRef] [PubMed]
- de Ceglia, M.; Decara, J.; Gaetani, S.; Rodríguez de Fonseca, F. Obesity as a Condition Determined by Food Addiction: Should Brain Endocannabinoid System Alterations Be the Cause and Its Modulation the Solution? Pharmaceuticals 2021, 14, 1002. [Google Scholar] [CrossRef] [PubMed]
- Bourdy, R.; Hertz, A.; Filliol, D.; Andry, V.; Goumon, Y.; Mendoza, J.; Olmstead, M.C.; Befort, K. The Endocannabinoid System Is Modulated in Reward and Homeostatic Brain Regions Following Diet-Induced Obesity in Rats: A Cluster Analysis Approach. Eur. J. Nutr. 2021, 60, 4621–4633. [Google Scholar] [CrossRef]
- Pucci, M.; Di Bonaventura, M.V.M.; Vezzoli, V.; Zaplatic, E.; Massimini, M.; Mai, S.; Sartorio, A.; Scacchi, M.; Persani, L.; Maccarrone, M.; et al. Preclinical and Clinical Evidence for a Distinct Regulation of Mu Opioid and Type 1 Cannabinoid Receptor Genes Expression in Obesity. Front. Genet. 2019, 10, 523. [Google Scholar] [CrossRef]
- Cifani, C.; Polidori, C.; Melotto, S.; Ciccocioppo, R.; Massi, M. A Preclinical Model of Binge Eating Elicited by Yo-Yo Dieting and Stressful Exposure to Food: Effect of Sibutramine, Fluoxetine, Topiramate, and Midazolam. Psychopharmacology (Berl) 2009, 204, 113–125. [Google Scholar] [CrossRef]
- Cifani, C.; Di Bonaventura, M.V.M.; Ciccocioppo, R.; Massi, M. Binge Eating in Female Rats Induced by Yo-Yo Dieting and Stress. In Animal Models of Eating Disorders; Humana Press: Totowa, NJ, USA, 2013; pp. 27–49. [Google Scholar]
- Zhang, S.; Zhornitsky, S.; Le, T.M.; Li, C.-S.R. Hypothalamic Responses to Cocaine and Food Cues in Individuals with Cocaine Dependence. Int. J. Neuropsychopharmacol. 2019, 22, 754–764. [Google Scholar] [CrossRef]
- Kania, A.; Szlaga, A.; Sambak, P.; Gugula, A.; Blasiak, E.; Micioni Di Bonaventura, M.V.; Hossain, M.A.; Cifani, C.; Hess, G.; Gundlach, A.L.; et al. RLN3/RXFP3 Signaling in the PVN Inhibits Magnocellular Neurons via M-like Current Activation and Contributes to Binge Eating Behavior. J. Neurosci. 2020, 40, 5362–5375. [Google Scholar] [CrossRef]
- Micioni Di Bonaventura, M.V.; Ubaldi, M.; Giusepponi, M.E.; Rice, K.C.; Massi, M.; Ciccocioppo, R.; Cifani, C. Hypothalamic CRF1 Receptor Mechanisms Are Not Sufficient to Account for Binge-like Palatable Food Consumption in Female Rats. Int. J. Eat. Disord. 2017, 50, 1194–1204. [Google Scholar] [CrossRef]
- Cifani, C.; Micioni Di Bonaventura, M.V.; Vitale, G.; Ruggieri, V.; Ciccocioppo, R.; Massi, M. Effect of Salidroside, Active Principle of Rhodiola Rosea Extract, on Binge Eating. Physiol. Behav. 2010, 101, 555–562. [Google Scholar] [CrossRef]
- Micioni Di Bonaventura, M.V.; Vitale, G.; Massi, M.; Cifani, C. Effect of Hypericum Perforatum Extract in an Experimental Model of Binge Eating in Female Rats. J. Obes. 2012, 2012, 956137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Micioni Di Bonaventura, M.V.; Ciccocioppo, R.; Romano, A.; Bossert, J.M.; Rice, K.C.; Ubaldi, M.; St Laurent, R.; Gaetani, S.; Massi, M.; Shaham, Y.; et al. Role of Bed Nucleus of the Stria Terminalis Corticotrophin-Releasing Factor Receptors in Frustration Stress-Induced Binge-like Palatable Food Consumption in Female Rats with a History of Food Restriction. J. Neurosci. 2014, 34, 11316–11324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, C.; Patterson, S.; Shah, K.; Longley, M.; Sirohi, S. Intermittent but Not Chronic Access to Western Diet Selectively Induced Binge-like Eating and Risk-taking Behavior in Rats. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Satta, V.; Scherma, M.; Piscitelli, F.; Usai, P.; Castelli, M.P.; Bisogno, T.; Fratta, W.; Fadda, P. Limited Access to a High Fat Diet Alters Endocannabinoid Tone in Female Rats. Front. Neurosci. 2018, 12, 40. [Google Scholar] [CrossRef]
- Micioni Di Bonaventura, M.V.; Micioni Di Bonaventura, E.; Polidori, C.; Cifani, C. Preclinical Models of Stress and Environmental Influences on Binge Eating. In Binge Eating; Springer International Publishing: Cham, Switerland, 2020; pp. 85–101. [Google Scholar]
- Micioni Di Bonaventura, M.V.; Micioni Di Bonaventura, E.; Botticelli, L.; Cifani, C. Impact of a History of Caloric Restriction and a Frustration Stress Manipulation on Binge-Like Eating Behavior in Female Rats: Preclinical Results. In Animal Models of Eating Disorders; Humana: New York, NY, USA, 2021; pp. 239–260. [Google Scholar]
- Brewerton, T.D.; Dansky, B.S.; Kilpatrick, D.G.; O’Neil, P.M. Which Comes First in the Pathogenesis of Bulimia Nervosa: Dieting or Bingeing? Int. J. Eat. Disord. 2000, 28, 259–264. [Google Scholar] [CrossRef]
- Crowther, J.H.; Sanftner, J.; Bonifazi, D.Z.; Shepherd, K.L. The Role of Daily Hassles in Binge Eating. Int. J. Eat. Disord. 2001, 29, 449–454. [Google Scholar] [CrossRef]
- Grilo, C.M.; Masheb, R.M.; Wilson, G.T. A Comparison of Different Methods for Assessing the Features of Eating Disorders in Patients with Binge Eating Disorder. J. Consult. Clin. Psychol. 2001, 69, 317–322. [Google Scholar] [CrossRef]
- Mitchell, J.E.; Hatsukami, D.; Eckert, E.D.; Pyle, R.L. Characteristics of 275 Patients with Bulimia. Am. J. Psychiatry 1985, 142, 482–485. [Google Scholar] [CrossRef]
- Polivy, J.; Herman, C.P. Dieting and Binging. A Causal Analysis. Am. Psychol. 1985, 40, 193–201. [Google Scholar] [CrossRef]
- Polivy, J.; Herman, C.P.; McFarlane, T. Effects of Anxiety on Eating: Does Palatability Moderate Distress-Induced Overeating in Dieters? J. Abnorm. Psychol. 1994, 103, 505–510. [Google Scholar] [CrossRef]
- Stice, E.; Agras, W.S.; Telch, C.F.; Halmi, K.A.; Mitchell, J.E.; Wilson, T. Subtyping Binge Eating-Disordered Women along Dieting and Negative Affect Dimensions. Int. J. Eat. Disord. 2001, 30, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Herman, C.P.; Polivy, J. From Dietary Restraint to Binge Eating: Attaching Causes to Effects. Appetite 1990, 14, 123–125; discussion 142–143. [Google Scholar] [CrossRef] [PubMed]
- Baucom, D.H.; Aiken, P.A. Effect of Depressed Mood in Eating among Obese and Nonobese Dieting and Nondieting Persons. J. Pers. Soc. Psychol. 1981, 41, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Crow, S.; Kendall, D.; Praus, B.; Thuras, P. Binge Eating and Other Psychopathology in Patients with Type II Diabetes Mellitus. Int. J. Eat. Disord. 2001, 30, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Greeno, C.G.; Wing, R.R. Stress-Induced Eating. Psychol. Bull. 1994, 115, 444–464. [Google Scholar] [CrossRef] [PubMed]
- Macht, M. Effects of High- and Low-Energy Meals on Hunger, Physiological Processes and Reactions to Emotional Stress. Appetite 1996, 26, 71–88. [Google Scholar] [CrossRef]
- Oliver, G.; Wardle, J. Perceived Effects of Stress on Food Choice. Physiol. Behav. 1999, 66, 511–515. [Google Scholar] [CrossRef]
- Polivy, J.; Herman, C.P. Distress and Eating: Why Do Dieters Overeat? Int. J. Eat. Disord. 1999, 26, 153–164. [Google Scholar] [CrossRef]
- Rutledge, T.; Linden, W. To Eat or Not to Eat: Affective and Physiological Mechanisms in the Stress-Eating Relationship. J. Behav. Med. 1998, 21, 221–240. [Google Scholar] [CrossRef]
- Wardle, J.; Steptoe, A.; Oliver, G.; Lipsey, Z. Stress, Dietary Restraint and Food Intake. J. Psychosom. Res. 2000, 48, 195–202. [Google Scholar] [CrossRef]
- Cardinal, P.; Bellocchio, L.; Clark, S.; Cannich, A.; Klugmann, M.; Lutz, B.; Marsicano, G.; Cota, D. Hypothalamic CB1 Cannabinoid Receptors Regulate Energy Balance in Mice. Endocrinology 2012, 153, 4136–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccioni, P.; Pes, D.; Carai, M.A.M.; Gessa, G.L.; Colombo, G. Suppression by the Cannabinoid CB1 Receptor Antagonist, Rimonabant, of the Reinforcing and Motivational Properties of a Chocolate-Flavoured Beverage in Rats. Behav. Pharmacol. 2008, 19, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Higgs, S.; Williams, C.M.; Kirkham, T.C. Cannabinoid Influences on Palatability: Microstructural Analysis of Sucrose Drinking after Delta(9)-Tetrahydrocannabinol, Anandamide, 2-Arachidonoyl Glycerol and SR141716. Psychopharmacology 2003, 165, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Pério, A.; Barnouin, M.C.; Poncelet, M.; Soubrié, P. Activity of SR141716 on Post-Reinforcement Pauses in Operant Responding for Sucrose Reward in Rats. Behav. Pharmacol. 2001, 12, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Cota, D.; Marsicano, G.; Tschöp, M.; Grübler, Y.; Flachskamm, C.; Schubert, M.; Auer, D.; Yassouridis, A.; Thöne-Reineke, C.; Ortmann, S.; et al. The Endogenous Cannabinoid System Affects Energy Balance via Central Orexigenic Drive and Peripheral Lipogenesis. J. Clin. Investig. 2003, 112, 423–431. [Google Scholar] [CrossRef]
- Marsicano, G.; Lutz, B. Expression of the Cannabinoid Receptor CB1 in Distinct Neuronal Subpopulations in the Adult Mouse Forebrain. Eur. J. Neurosci. 1999, 11, 4213–4225. [Google Scholar] [CrossRef]
- Mazier, W.; Saucisse, N.; Gatta-Cherifi, B.; Cota, D. The Endocannabinoid System: Pivotal Orchestrator of Obesity and Metabolic Disease. Trends Endocrinol. Metab. 2015, 26, 524–537. [Google Scholar] [CrossRef]
- Fernández-Ruiz, J.J.; Muñoz, R.M.; Romero, J.; Villanua, M.A.; Makriyannis, A.; Ramos, J.A. Time Course of the Effects of Different Cannabimimetics on Prolactin and Gonadotrophin Secretion: Evidence for the Presence of CB1 Receptors in Hypothalamic Structures and Their Involvement in the Effects of Cannabimimetics. Biochem. Pharmacol. 1997, 53, 1919–1927. [Google Scholar] [CrossRef]
- Jamshidi, N.; Taylor, D.A. Anandamide Administration into the Ventromedial Hypothalamus Stimulates Appetite in Rats. Br. J. Pharmacol. 2001, 134, 1151–1154. [Google Scholar] [CrossRef] [Green Version]
- Soria-Gómez, E.; Massa, F.; Bellocchio, L.; Rueda-Orozco, P.E.; Ciofi, P.; Cota, D.; Oliet, S.H.R.; Prospéro-García, O.; Marsicano, G. Cannabinoid Type-1 Receptors in the Paraventricular Nucleus of the Hypothalamus Inhibit Stimulated Food Intake. Neuroscience 2014, 263, 46–53. [Google Scholar] [CrossRef]
- Williams, C.M.; Kirkham, T.C. Anandamide Induces Overeating: Mediation by Central Cannabinoid (CB1) Receptors. Psychopharmacology 1999, 143, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Sanchis-Segura, C.; Cline, B.H.; Marsicano, G.; Lutz, B.; Spanagel, R. Reduced Sensitivity to Reward in CB1 Knockout Mice. Psychopharmacology 2004, 176, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Trojniar, W.; Wise, R.A. Facilitory Effect of Delta 9-Tetrahydrocannabinol on Hypothalamically Induced Feeding. Psychopharmacology 1991, 103, 172–176. [Google Scholar] [CrossRef]
- Piazza, P.V.; Cota, D.; Marsicano, G. The CB1 Receptor as the Cornerstone of Exostasis. Neuron 2017, 93, 1252–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berridge, K.C.; Valenstein, E.S. What Psychological Process Mediates Feeding Evoked by Electrical Stimulation of the Lateral Hypothalamus? Behav. Neurosci. 1991, 105, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Hanus, L.; Avraham, Y.; Ben-Shushan, D.; Zolotarev, O.; Berry, E.M.; Mechoulam, R. Short-Term Fasting and Prolonged Semistarvation Have Opposite Effects on 2-AG Levels in Mouse Brain. Brain Res. 2003, 983, 144–151. [Google Scholar] [CrossRef]
- Kirkham, T.C.; Williams, C.M.; Fezza, F.; Di Marzo, V. Endocannabinoid Levels in Rat Limbic Forebrain and Hypothalamus in Relation to Fasting, Feeding and Satiation: Stimulation of Eating by 2-Arachidonoyl Glycerol. Br. J. Pharmacol. 2002, 136, 550–557. [Google Scholar] [CrossRef] [Green Version]
- Di Marzo, V.; Goparaju, S.K.; Wang, L.; Liu, J.; Bátkai, S.; Járai, Z.; Fezza, F.; Miura, G.I.; Palmiter, R.D.; Sugiura, T.; et al. Leptin-Regulated Endocannabinoids Are Involved in Maintaining Food Intake. Nature 2001, 410, 822–825. [Google Scholar] [CrossRef]
- Monteleone, P.; Matias, I.; Martiadis, V.; De Petrocellis, L.; Maj, M.; Di Marzo, V. Blood Levels of the Endocannabinoid Anandamide Are Increased in Anorexia Nervosa and in Binge-Eating Disorder, but Not in Bulimia Nervosa. Neuropsychopharmacology 2005, 30, 1216–1221. [Google Scholar] [CrossRef] [Green Version]
- Gatta-Cherifi, B.; Matias, I.; Vallée, M.; Tabarin, A.; Marsicano, G.; Piazza, P.V.; Cota, D. Simultaneous Postprandial Deregulation of the Orexigenic Endocannabinoid Anandamide and the Anorexigenic Peptide YY in Obesity. Int. J. Obes. (Lond.) 2012, 36, 880–885. [Google Scholar] [CrossRef]
- Soria-Gómez, E.; Matias, I.; Rueda-Orozco, P.E.; Cisneros, M.; Petrosino, S.; Navarro, L.; Di Marzo, V.; Prospéro-García, O. Pharmacological Enhancement of the Endocannabinoid System in the Nucleus Accumbens Shell Stimulates Food Intake and Increases C-Fos Expression in the Hypothalamus. Br. J. Pharmacol. 2007, 151, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Touriño, C.; Oveisi, F.; Lockney, J.; Piomelli, D.; Maldonado, R. FAAH Deficiency Promotes Energy Storage and Enhances the Motivation for Food. Int. J. Obes. (Lond.) 2010, 34, 557–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Addario, C.; Di Francesco, A.; Arosio, B.; Gussago, C.; Dell’Osso, B.; Bari, M.; Galimberti, D.; Scarpini, E.; Altamura, A.C.; Mari, D.; et al. Epigenetic Regulation of Fatty Acid Amide Hydrolase in Alzheimer Disease. PLoS ONE 2012, 7, e39186. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Hutchison, K.E.; Bryan, A.D.; Filbey, F.M.; Calhoun, V.D.; Claus, E.D.; Lin, D.; Sui, J.; Du, Y.; Liu, J. Opposite Epigenetic Associations With Alcohol Use and Exercise Intervention. Front. Psychiatry 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Keski-Rahkonen, A. Epidemiology of Binge Eating Disorder: Prevalence, Course, Comorbidity, and Risk Factors. Curr. Opin. Psychiatry 2021, 34, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Romano, A.; Micioni Di Bonaventura, M.V.; Gallelli, C.A.; Koczwara, J.B.; Smeets, D.; Giusepponi, M.E.; De Ceglia, M.; Friuli, M.; Micioni Di Bonaventura, E.; Scuderi, C.; et al. Oleoylethanolamide Decreases Frustration Stress-Induced Binge-like Eating in Female Rats: A Novel Potential Treatment for Binge Eating Disorder. Neuropsychopharmacology 2020, 45, 1931–1941. [Google Scholar] [CrossRef]
- Del Bello, F.; Micioni Di Bonaventura, M.V.; Bonifazi, A.; Wünsch, B.; Schepmann, D.; Giancola, J.B.; Micioni Di Bonaventura, E.; Vistoli, G.; Giorgioni, G.; Quaglia, W.; et al. Investigation of the Role of Chirality in the Interaction with σ Receptors and Effect on Binge Eating Episode of a Potent Σ1 Antagonist Analogue of Spipethiane. ACS Chem. Neurosci. 2019, 10, 3391–3397. [Google Scholar] [CrossRef]
- Cifani, C.; Micioni Di Bonaventura, E.; Botticelli, L.; Del Bello, F.; Giorgioni, G.; Pavletić, P.; Piergentili, A.; Quaglia, W.; Bonifazi, A.; Schepmann, D.; et al. Novel Highly Potent and Selective Sigma1 Receptor Antagonists Effectively Block the Binge Eating Episode in Female Rats. ACS Chem. Neurosci. 2020, 11, 3107–3116. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction: Twenty-Something Years On. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- Dahl, J.A.; Collas, P. Q2ChIP, a Quick and Quantitative Chromatin Immunoprecipitation Assay, Unravels Epigenetic Dynamics of Developmentally Regulated Genes in Human Carcinoma Cells. Stem Cells 2007, 25, 1037–1046. [Google Scholar] [CrossRef]
- Caputi, F.F.; Di Benedetto, M.; Carretta, D.; Bastias del Carmen Candia, S.; D’Addario, C.; Cavina, C.; Candeletti, S.; Romualdi, P. Dynorphin/KOP and Nociceptin/NOP Gene Expression and Epigenetic Changes by Cocaine in Rat Striatum and Nucleus Accumbens. Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 49, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Pucci, M.; D’Addario, C. Assessing Gene Expression of the Endocannabinoid System. In Endocannabinoid Signaling; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2016; Volume 1412, pp. 237–246. [Google Scholar]
- Cifani, C.; Micioni Di Bonaventura, M.V.; Pucci, M.; Giusepponi, M.E.; Romano, A.; Di Francesco, A.; Maccarrone, M.; D’Addario, C. Regulation of Hypothalamic Neuropeptides Gene Expression in Diet Induced Obesity Resistant Rats: Possible Targets for Obesity Prediction? Front. Neurosci. 2015, 9, 187. [Google Scholar] [CrossRef] [Green Version]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.R.; Bock, M. An Extended ΔCT-Method Facilitating Normalisation with Multiple Reference Genes Suited for Quantitative RT-PCR Analyses of Human Hepatocyte-Like Cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Reed, K.; Poulin, M.L.; Yan, L.; Parissenti, A.M. Comparison of Bisulfite Sequencing PCR with Pyrosequencing for Measuring Differences in DNA Methylation. Anal. Biochem. 2010, 397, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Šestáková, Š.; Šálek, C.; Remešová, H. DNA Methylation Validation Methods: A Coherent Review with Practical Comparison. Biol. Proced. Online 2019, 21, 19. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef] [Green Version]
- Sergi, M.; Battista, N.; Montesano, C.; Curini, R.; Maccarrone, M.; Compagnone, D. Determination of the Two Major Endocannabinoids in Human Plasma by μ-SPE Followed by HPLC-MS/MS. Anal. Bioanal. Chem. 2013, 405, 785–793. [Google Scholar] [CrossRef]
- Alboni, S.; Micioni Di Bonaventura, M.V.; Benatti, C.; Giusepponi, M.E.; Brunello, N.; Cifani, C. Hypothalamic Expression of Inflammatory Mediators in an Animal Model of Binge Eating. Behav. Brain Res. 2017, 320, 420–430. [Google Scholar] [CrossRef]
- Micioni Di Bonaventura, M.V.; Lutz, T.A.; Romano, A.; Pucci, M.; Geary, N.; Asarian, L.; Cifani, C. Estrogenic Suppression of Binge-like Eating Elicited by Cyclic Food Restriction and Frustrative-Nonreward Stress in Female Rats. Int. J. Eat. Disord. 2017, 50, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Culbert, K.M.; Racine, S.E.; Klump, K.L. Hormonal Factors and Disturbances in Eating Disorders. Curr. Psychiatry Rep. 2016, 18, 65. [Google Scholar] [CrossRef]
- de Ávila, C.; Chometton, S.; Calvez, J.; Guèvremont, G.; Kania, A.; Torz, L.; Lenglos, C.; Blasiak, A.; Rosenkilde, M.M.; Holst, B.; et al. Estrous Cycle Modulation of Feeding and Relaxin-3/Rxfp3 MRNA Expression: Implications for Estradiol Action. Neuroendocrinology 2021, 111, 1201–1218. [Google Scholar] [CrossRef] [PubMed]
- Edler, C.; Lipson, S.F.; Keel, P.K. Ovarian Hormones and Binge Eating in Bulimia Nervosa. Psychol. Med. 2007, 37, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Klump, K.L.; Culbert, K.M.; Sisk, C.L. Sex Differences in Binge Eating: Gonadal Hormone Effects Across Development. Annu. Rev. Clin. Psychol. 2017, 13, 183–207. [Google Scholar] [CrossRef]
- Lamontagne, S.J.; Wilkin, M.M.; Menard, J.L.; Olmstead, M.C. Mid-Adolescent Stress Differentially Affects Binge-like Intake of Sucrose across Estrous Cycles in Female Rats. Physiol. Behav. 2021, 228, 113194. [Google Scholar] [CrossRef] [PubMed]
- Marcondes, F.K.; Bianchi, F.J.; Tanno, A.P. Determination of the Estrous Cycle Phases of Rats: Some Helpful Considerations. Braz. J. Biol. 2002, 62, 609–614. [Google Scholar] [CrossRef] [Green Version]
- Freeman, M. Neuroendocrine Control of the Ovarian Cycle of the Rat. In Physiology of Reproduction, 2nd ed.; Knobil, E., Neill, E., Eds.; Raven Press: New York, NY, USA, 1994; Volume 45, pp. 613–658. [Google Scholar]
- Nequin, L.G.; Alvarez, J.; Schwartz, N.B. Measurement of Serum Steroid and Gonadotropin Levels and Uterine and Ovarian Variables throughout 4 Day and 5 Day Estrous Cycles in the Rat. Biol. Reprod. 1979, 20, 659–670. [Google Scholar] [CrossRef] [Green Version]
- Marashi, V.; Rülicke, T. The Bruce Effect in Norway Rats. Biol. Reprod. 2012, 86, 1–5. [Google Scholar] [CrossRef]
- Sharp, P.; Villano, J.S. The Laboratory Rat; CRC Press: Boca Raton, FL, USA, 2012; ISBN 9780429152405. [Google Scholar]
- Lohmiller, J.J.; Swing, S.P.; Hanson, M.M. Reproduction and Breeding. In The Laboratory Rat, 3rd ed.; Suckow, M.A., Hankenson, F.C., Wilson, R.P., Eds.; Academic Press: London, UK, 2020; pp. 157–179. [Google Scholar]
- Shirley, B. Partial Synchrony of the Oestrous Cycles of Rats Introduced to a New Environment. J. Endocrinol. 1978, 77, 195–202. [Google Scholar] [CrossRef]
- Schank, J.C. Do Norway Rats (Rattus Norvegicus) Synchronize Their Estrous Cycles? Physiol. Behav. 2001, 72, 129–139. [Google Scholar] [CrossRef]
- Howland, B.E. Effect of Restricted Feed Intake on LH Levels in Female Rats. J. Anim. Sci. 1972, 34, 445–447. [Google Scholar] [CrossRef] [PubMed]
- Knuth, U.A.; Friesen, H.G. Starvation Induced Anoestrus: Effect of Chronic Food Restriction on Body Weight, Its Influence on Oestrous Cycle and Gonadotrophin Secretion in Rats. Acta Endocrinol. (Cph.) 1983, 104, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Lintern-Moore, S.; Everitt, A.V. The Effect of Restricted Food Intake on the Size and Composition of the Ovarian Follicle Population in the Wistar Rat. Biol. Reprod. 1978, 19, 688–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, M.; Yamaguchi, K.; Marumo, H.; Imai, K. Effects of Food Restriction on Reproductive and Toxicological Parameters in Rats--in Search of Suitable Feeding Regimen in Long-Term Tests. J. Toxicol. Sci. 1997, 22, 427–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tropp, J.; Markus, E.J. Effects of Mild Food Deprivation on the Estrous Cycle of Rats. Physiol. Behav. 2001, 73, 553–559. [Google Scholar] [CrossRef] [PubMed]




| Feeding Tests | NR + NS | NR + S | R + NS | R + S | ||||
|---|---|---|---|---|---|---|---|---|
| 15 min | 120 min | 15 min | 120 min | 15 min | 120 min | 15 min | 120 min | |
| FIRST | 84.1 ± 5.6 | 113.2 ± 9.5 | 70.6 ± 6.8 | 103.0 ± 12.7 | 87.1 ± 7.4 | 118.5 ± 8.0 | 131.3 ± 7.1 ** | 177.8 ± 4.4 ** |
| SECOND | 82.3 ± 7.4 | 120.7 ± 10.2 | 72.5 ± 3.0 | 101.3 ± 5.9 | 74.6 ± 5.2 | 117.4 ± 6.5 | 127.5 ± 4.5 ** | 166.1 ± 10.0 ** |
| THIRD | 73.1 ± 5.8 | 105.7 ± 7.8 | 78.9 ± 5.6 | 107.9 ± 5.5 | 87.0 ± 4.8 | 116.3 ± 5.0 | 132.2 ± 10.0 ** | 179.4 ± 14.4 ** |
| FOURTH | 72.3 ± 3.9 | 107.3 ± 1.0 | 83.6 ± 4.6 | 113.7 ± 3.1 | 86.6 ± 3.9 | 109.7 ± 3.6 | 130.7 ± 3.6 ** | 167.0 ± 9.6 ** |
| % DNA Methylation | Relative Gene Expression | |||
|---|---|---|---|---|
| Spearman’s r | p Value | Equation | p Value | |
| Faah CpG site 4 | −0.4404 | 0.059 | Y = −2.853 × X + 7.706 | 0.0662 |
| Faah CpG site 5 | −0.5634 | 0.0120 | Y = −0.7290 × X + 1.643 | 0.0179 |
| Dagl CpG site 1 | −0.258 | 0.301 | Y = −0.7624 × X + 3.216 | 0.4919 |
| Magl CpG site 4 | −0.1685 | 0.5300 | Y = −0.6307 × X + 3.654 | 0.3297 |
| Magl CpG site 6 | −0.1694 | 0.5121 | Y = −0.2996 × X + 1.994 | 0.2883 |
| AEA levels | ||||
| % DNA Methylation | Spearman’s r | p value | Equation | p value |
| Faah CpG site 4 | −0.3561 | 0.1345 | Y = −23.47 × X + 6.111 | 0.0826 |
| Faah CpG site 5 | −0.5353 | 0.0182 | Y = −4.495 × X + 1.179 | 0.1065 |
| Relative gene expression | ||||
| Faah | 0.4842 | 0.0305 | Y = 0.05542 × X − 0.01095 | 0.0288 |
| Cnr1 | 0.4055 | 0.0761 | Y = 0.02809 × X + 0.003122 | 0.1044 |
| Relative gene expression | Relative gene expression Faah | |||
| Spearman’s r | p value | Equation | p value | |
| Cnr1 | 0.3796 | 0.0987 | Y = 0.2759 × X + 0.5403 | 0.0678 |
| AEA levels | Palatable food intake (kcal/kg) | |||
| Spearman’s r | p value | Equation | p value | |
| 0.6045 | 0.0048 | Y = 535.0 × X + 107.8 | 0.0013 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pucci, M.; D’Addario, C.; Micioni Di Bonaventura, E.; Mercante, F.; Annunzi, E.; Fanti, F.; Sergi, M.; Botticelli, L.; Einaudi, G.; Cifani, C.; et al. Endocannabinoid System Regulation in Female Rats with Recurrent Episodes of Binge Eating. Int. J. Mol. Sci. 2022, 23, 15228. https://doi.org/10.3390/ijms232315228
Pucci M, D’Addario C, Micioni Di Bonaventura E, Mercante F, Annunzi E, Fanti F, Sergi M, Botticelli L, Einaudi G, Cifani C, et al. Endocannabinoid System Regulation in Female Rats with Recurrent Episodes of Binge Eating. International Journal of Molecular Sciences. 2022; 23(23):15228. https://doi.org/10.3390/ijms232315228
Chicago/Turabian StylePucci, Mariangela, Claudio D’Addario, Emanuela Micioni Di Bonaventura, Francesca Mercante, Eugenia Annunzi, Federico Fanti, Manuel Sergi, Luca Botticelli, Giacomo Einaudi, Carlo Cifani, and et al. 2022. "Endocannabinoid System Regulation in Female Rats with Recurrent Episodes of Binge Eating" International Journal of Molecular Sciences 23, no. 23: 15228. https://doi.org/10.3390/ijms232315228






