Meta-Analysis of Heat-Stressed Transcriptomes Using the Public Gene Expression Database from Human and Mouse Samples
Abstract
:1. Introduction
2. Results
2.1. Overview of Analysis Scheme
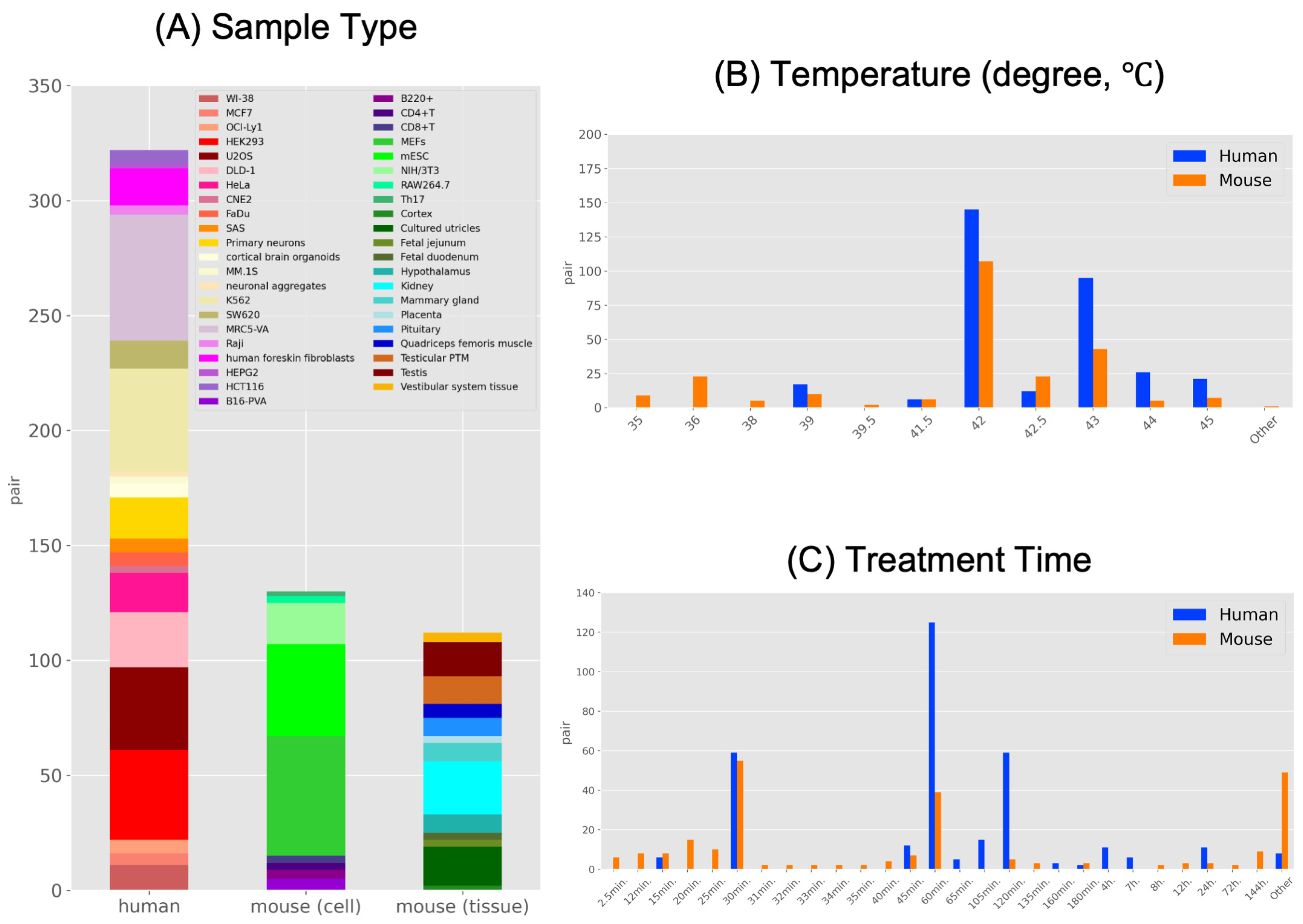
2.2. Characteristics of HS-Related High-Throughput Sequencing Data
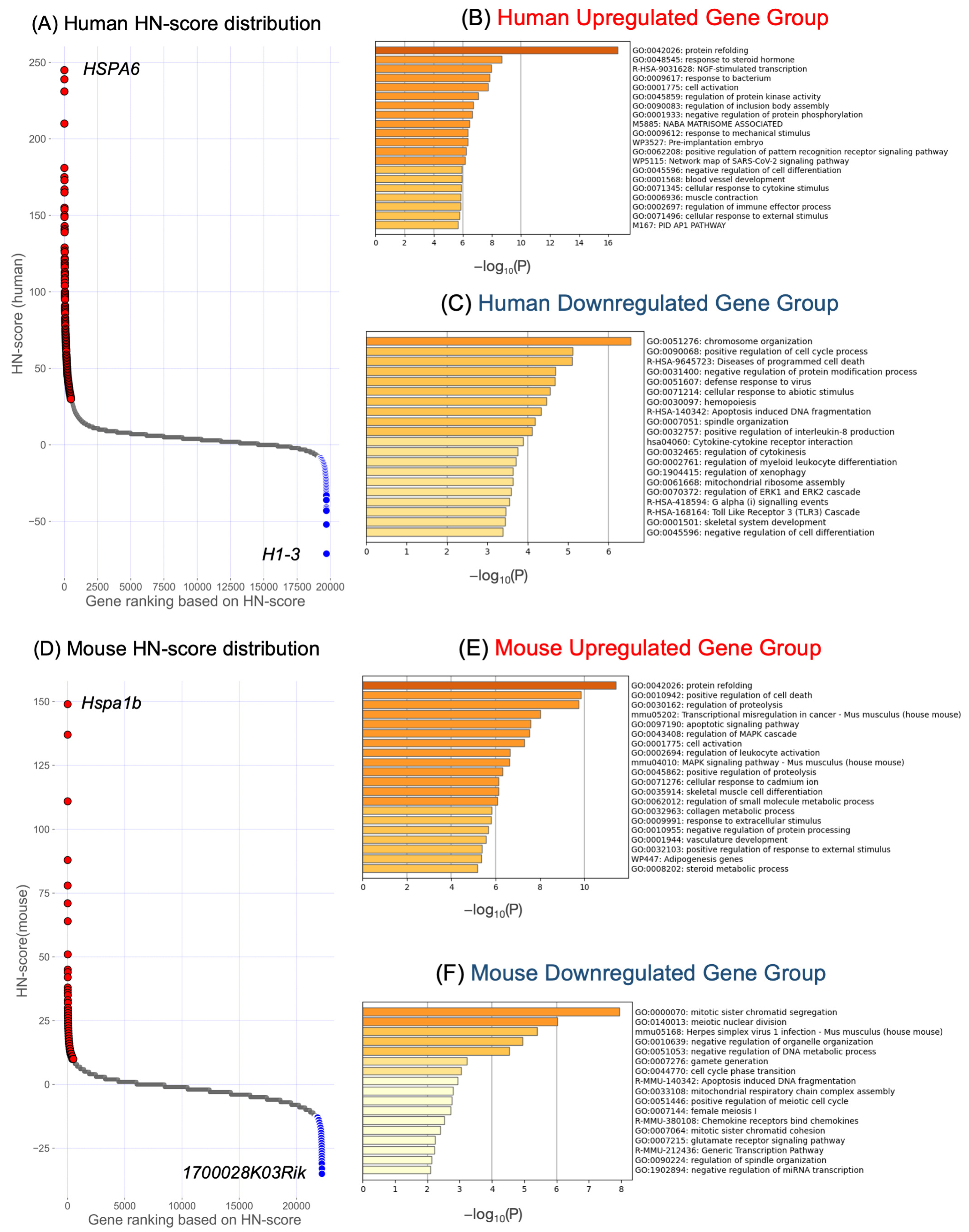
2.3. Verification of Upregulated and Downregulated Genes in Each Organism
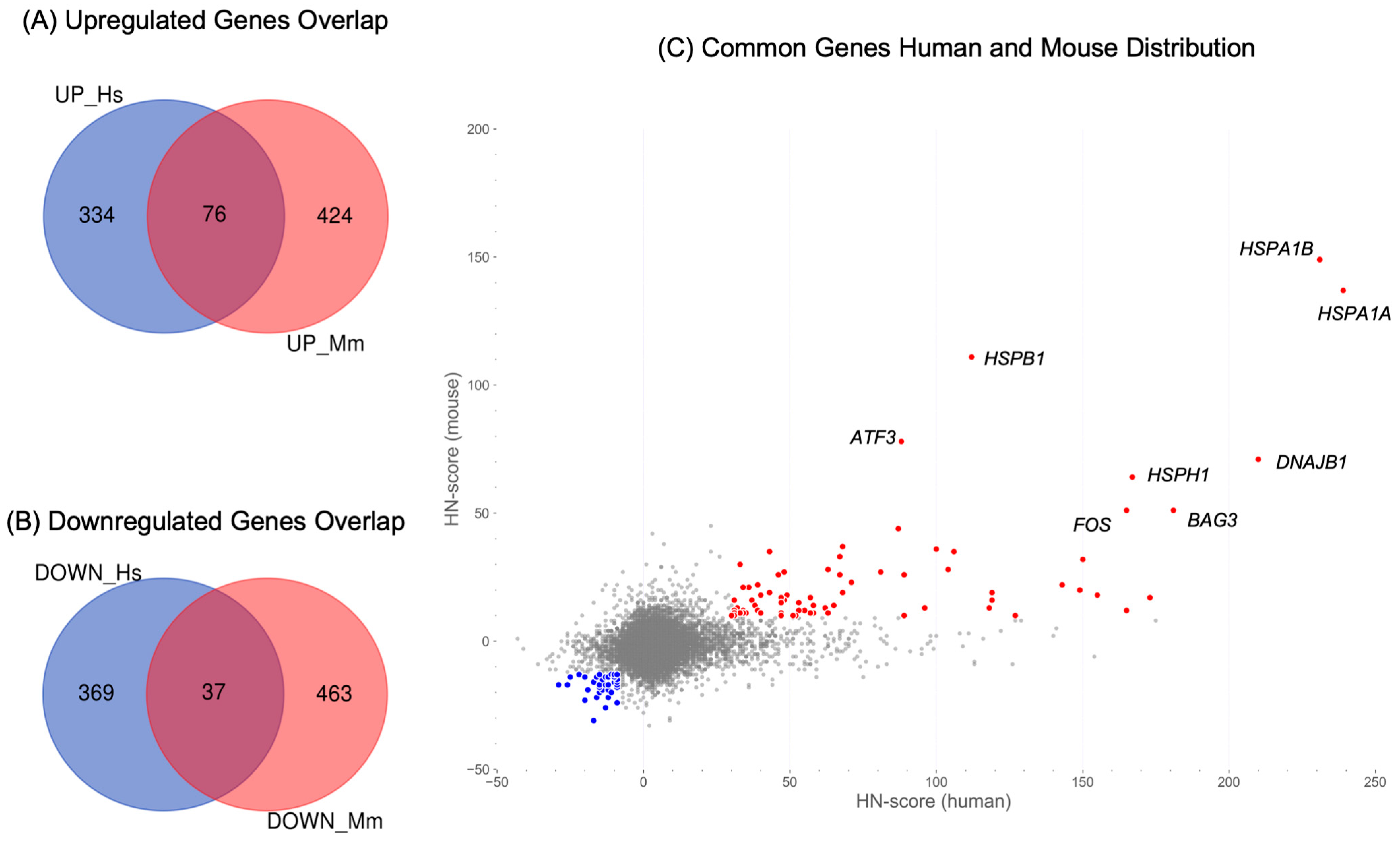
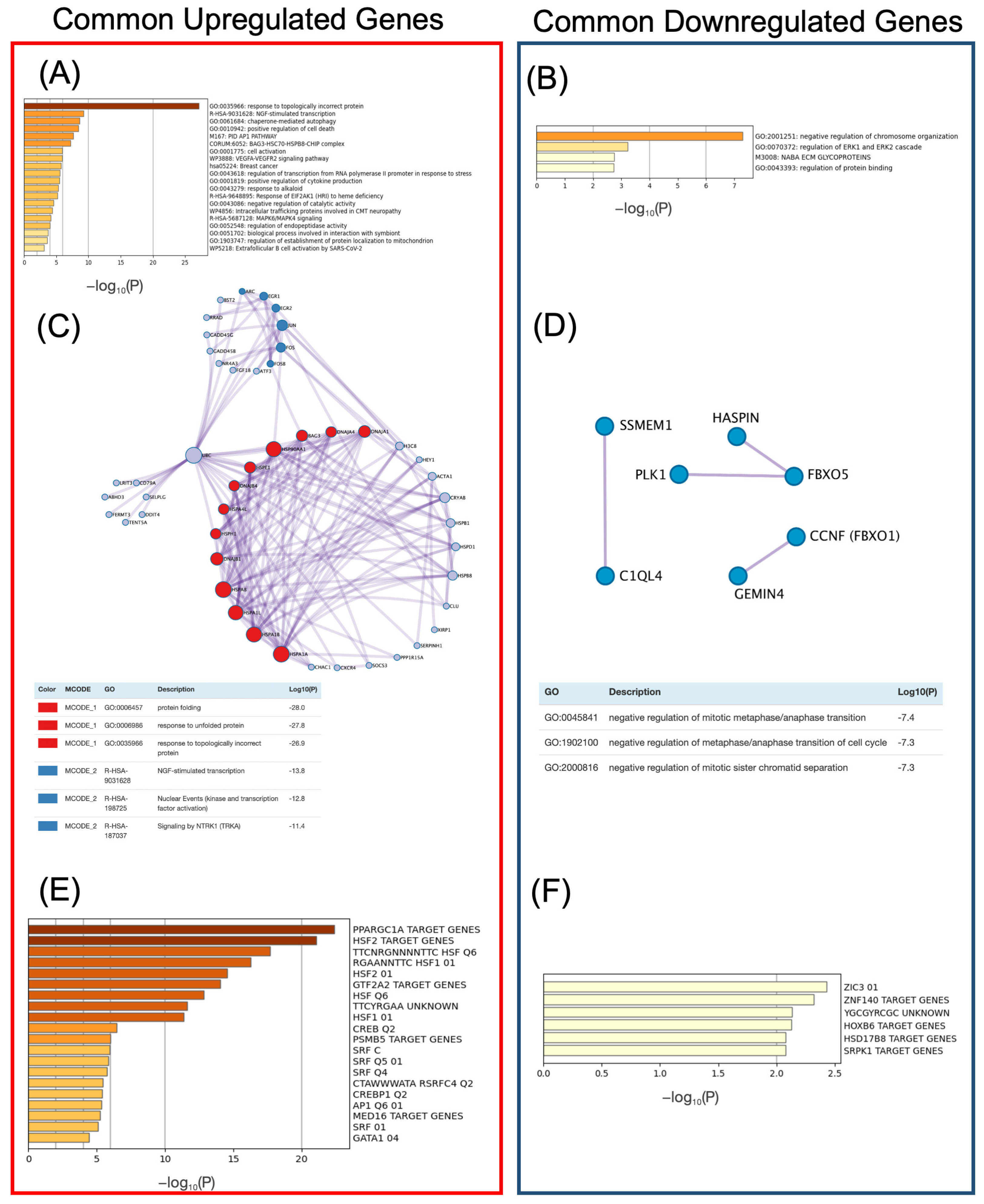
2.4. Identification of Common Upregulated or Downregulated Genes in Human and Mouse
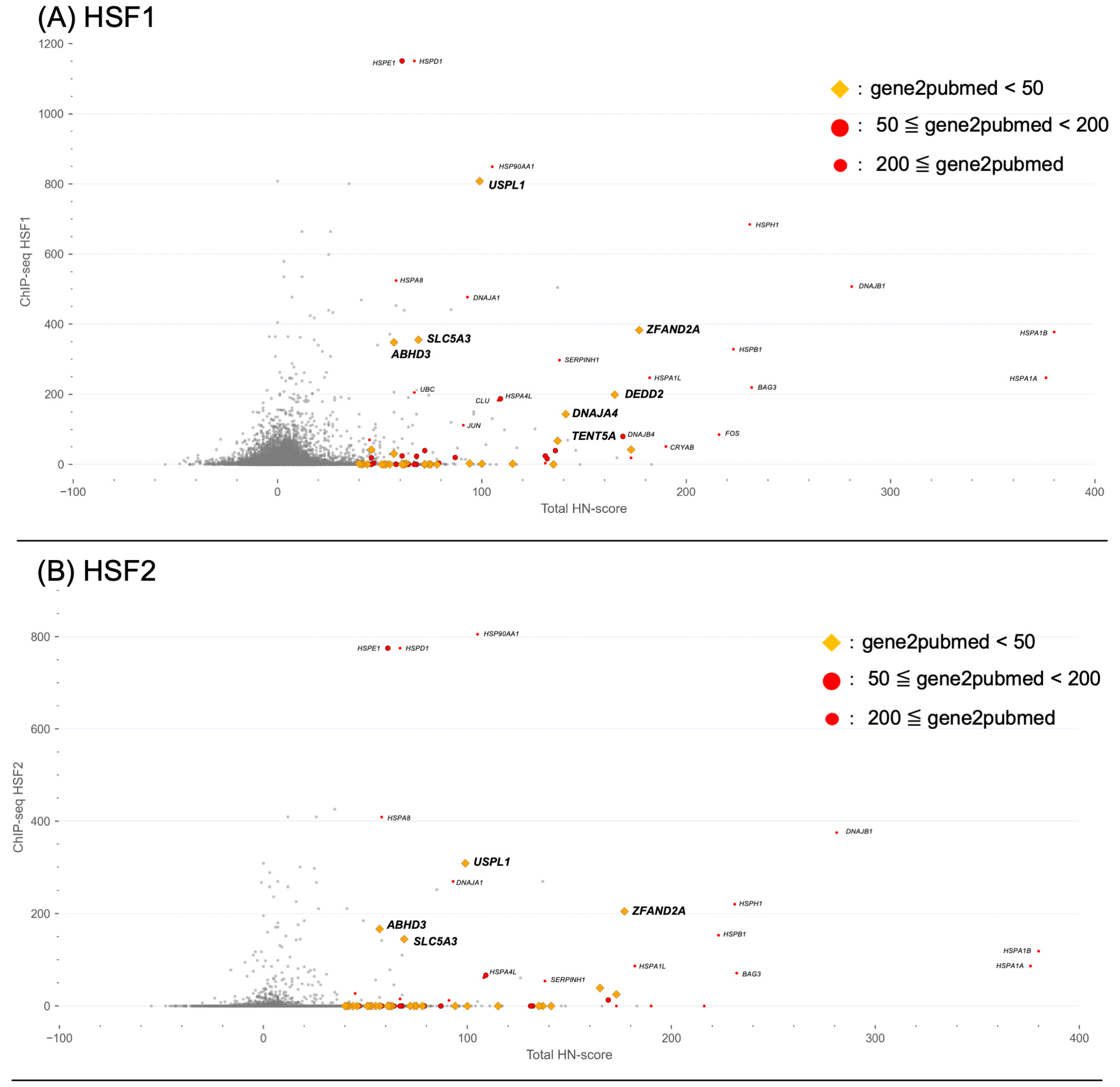
2.5. Integration of HN-Score, Transcription Factor-Binding Information, and Literature Information
3. Discussion
4. Materials and Methods
4.1. Curation of Gene Expression Data from Public Database
4.2. Quantification of Gene Expression Data Using the Analysis Pipeline
4.3. Calculation of HN-Ratio
4.4. Classification of Genes Based on HN-Ratio
4.5. Calculation of HN-Score
4.6. Analysis of Selected Gene Sets in Human and Mouse
4.7. Comprehensive Analysis of Common Upregulated Genes in Human and Mouse
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Romanello, M.; Di Napoli, C.; Drummond, P.; Green, C.; Kennard, H.; Lampard, P.; Scamman, D.; Arnell, N.; Ayeb-Karlsson, S.; Ford, L.B.; et al. The 2022 report of the Lancet Countdown on health and climate change: Health at the mercy of fossil fuels. Lancet 2022, 400, 1619–1654. [Google Scholar] [CrossRef]
- Perkins-Kirkpatrick, S.E.; Lewis, S.C. Increasing trends in regional heatwaves. Nat. Commun. 2020, 11, 3357. [Google Scholar] [CrossRef]
- Székely, M.; Carletto, L.; Garami, A. The pathophysiology of heat exposure. Temperature 2015, 2, 452. [Google Scholar] [CrossRef]
- Velichko, A.K.; Markova, E.N.; Petrova, N.V.; Razin, S.V.; Kantidze, O.L. Mechanisms of heat shock response in mammals. Cell. Mol. Life Sci. 2013, 70, 4229–4241. [Google Scholar] [CrossRef]
- Qian, L.; Song, X.; Ren, H.; Gong, J.; Cheng, S. Mitochondrial mechanism of heat stress-induced injury in rat cardiomyocyte. Cell Stress Chaperones 2004, 9, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Zukiene, R.; Nauciene, Z.; Ciapaite, J.; Mildaziene, V. Acute temperature resistance threshold in heart mitochondria: Febrile temperature activates function but exceeding it collapses the membrane barrier. Int. J. Hyperth. 2010, 26, 56–66. [Google Scholar] [CrossRef]
- Balogh, G.; Maulucci, G.; Gombos, I.; Horváth, I.; Török, Z.; Péter, M.; Fodor, E.; Páli, T.; Benkő, S.; Parasassi, T.; et al. Heat Stress Causes Spatially-Distinct Membrane Re-Modelling in K562 Leukemia Cells. PLoS ONE 2011, 6, e21182. [Google Scholar] [CrossRef]
- Habibi, P.; Ostad, S.N.; Heydari, A.; Aliebrahimi, S.; Montazeri, V.; Foroushani, A.R.; Monazzam, M.R.; Ghazi-Khansari, M.; Golbabaei, F. Effect of heat stress on DNA damage: A systematic literature review. Int. J. Biometeorol. 2022, 66, 2147–2158. [Google Scholar] [CrossRef]
- Kantidze, O.L.; Velichko, A.K.; Luzhin, A.V.; Razin, S.V. Heat Stress-Induced DNA Damage. Acta Nat. 2016, 8, 75–78. Available online: https://www.ncbi.nlm.nih.gov/pubmed/27437141 (accessed on 1 August 2023). [CrossRef]
- Bruskov, V.I.; Malakhova, L.V.; Masalimov, Z.K.; Chernikov, A.V. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 2002, 30, 1354–1363. [Google Scholar] [CrossRef]
- Lepock, J.R. How do cells respond to their thermal environment? Int. J. Hyperth. 2005, 21, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Bono, H.; Hirota, K. Meta-Analysis of Hypoxic Transcriptomes from Public Databases. Biomedicines 2020, 8, 10. [Google Scholar] [CrossRef]
- Ono, Y.; Bono, H. Multi-Omic Meta-Analysis of Transcriptomes and the Bibliome Uncovers Novel Hypoxia-Inducible Genes. Biomedicines 2021, 9, 582. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Ono, Y.; Bono, H. Comparison of Oxidative and Hypoxic Stress Responsive Genes from Meta-Analysis of Public Transcriptomes. Biomedicines 2021, 9, 1830. [Google Scholar] [CrossRef]
- Meta-analysis in basic biology. Nat. Methods 2016, 13, 959. [CrossRef]
- Tamura, K.; Bono, H. Meta-Analysis of RNA Sequencing Data of Arabidopsis and Rice under Hypoxia. Life 2022, 12, 1079. [Google Scholar] [CrossRef]
- Toga, K.; Yokoi, K.; Bono, H. Meta-Analysis of Transcriptomes in Insects Showing Density-Dependent Polyphenism. Insects 2022, 13, 864. [Google Scholar] [CrossRef]
- Yonezawa, S. Meta-Analysis of Heat-stressed Gene Expression Data in Human and Mouse. Figshare 2023. [Google Scholar] [CrossRef]
- Yu, H.; Kohki, Y.; Ryuichiro, Y.; Yusuke, K.; Ryoko, K.; Yasunari, M.; Kaito, I.; Yuto, U.; Haruka, H.; Yasumizu, Y. yyoshiaki/ikra: ikra v2.0. Zenodo 2021. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Eppig, J.T. Mouse Genome Informatics (MGI) Resource: Genetic, Genomic, and Biological Knowledgebase for the Laboratory Mouse. ILAR J. 2017, 58, 17–41. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Oki, S.; Ohta, T.; Shioi, G.; Hatanaka, H.; Ogasawara, O.; Okuda, Y.; Kawaji, H.; Nakaki, R.; Sese, J.; Meno, C. ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018, 19, e46255. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Drabløs, F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016, 62, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Sandqvist, A.; Björk, J.K.; Åkerfelt, M.; Chitikova, Z.; Grichine, A.; Vourc’H, C.; Jolly, C.; Salminen, T.A.; Nymalm, Y.; Sistonen, L. Heterotrimerization of Heat-Shock Factors 1 and 2 Provides a Transcriptional Switch in Response to Distinct Stimuli. Mol. Biol. Cell 2009, 20, 1340–1347. [Google Scholar] [CrossRef]
- Xu, L.; Ma, X.; Bagattin, A.; Mueller, E. The transcriptional coactivator PGC1α protects against hyperthermic stress via cooperation with the heat shock factor HSF1. Cell Death Dis. 2016, 7, e2102. [Google Scholar] [CrossRef] [PubMed]
- Long, J.Z.; Cisar, J.S.; Milliken, D.; Niessen, S.; Wang, C.; Trauger, S.A.; Siuzdak, G.; Cravatt, B.F. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat. Chem. Biol. 2011, 7, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Trotta, E.; Brandi, R.; Arisi, I.; Coccia, M.; Santoro, M.G. AIRAP, a New Human Heat Shock Gene Regulated by Heat Shock Factor 1. J. Biol. Chem. 2010, 285, 13607–13615. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Chachami, G.; Kozaczkiewicz, L.; Winter, U.; Stankovic-Valentin, N.; Haas, P.; Hofmann, K.; Urlaub, H.; Ovaa, H.; Wittbrodt, J.; et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep. 2012, 13, 930–938. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Download SRA Sequences from Entrez Search Results. Available online: https://www.ncbi.nlm.nih.gov/sra/docs/sradownload/ (accessed on 30 March 2023).
- Babraham Bioinformatics-Trim Galore! Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 5 April 2023).
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Bono, H. Meta-Analysis of Oxidative Transcriptomes in Insects. Antioxidants 2021, 10, 345. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yonezawa, S.; Bono, H. Meta-Analysis of Heat-Stressed Transcriptomes Using the Public Gene Expression Database from Human and Mouse Samples. Int. J. Mol. Sci. 2023, 24, 13444. https://doi.org/10.3390/ijms241713444
Yonezawa S, Bono H. Meta-Analysis of Heat-Stressed Transcriptomes Using the Public Gene Expression Database from Human and Mouse Samples. International Journal of Molecular Sciences. 2023; 24(17):13444. https://doi.org/10.3390/ijms241713444
Chicago/Turabian StyleYonezawa, Sora, and Hidemasa Bono. 2023. "Meta-Analysis of Heat-Stressed Transcriptomes Using the Public Gene Expression Database from Human and Mouse Samples" International Journal of Molecular Sciences 24, no. 17: 13444. https://doi.org/10.3390/ijms241713444





