RpoS-Regulated Genes and Phenotypes in the Phytopathogenic Bacterium Pectobacterium atrosepticum
Abstract
:1. Introduction
2. Results and Discussion
2.1. Phenotype of the P. atrosepticum RpoS Mutant
2.1.1. Growth and Primary Stress (Starvation)
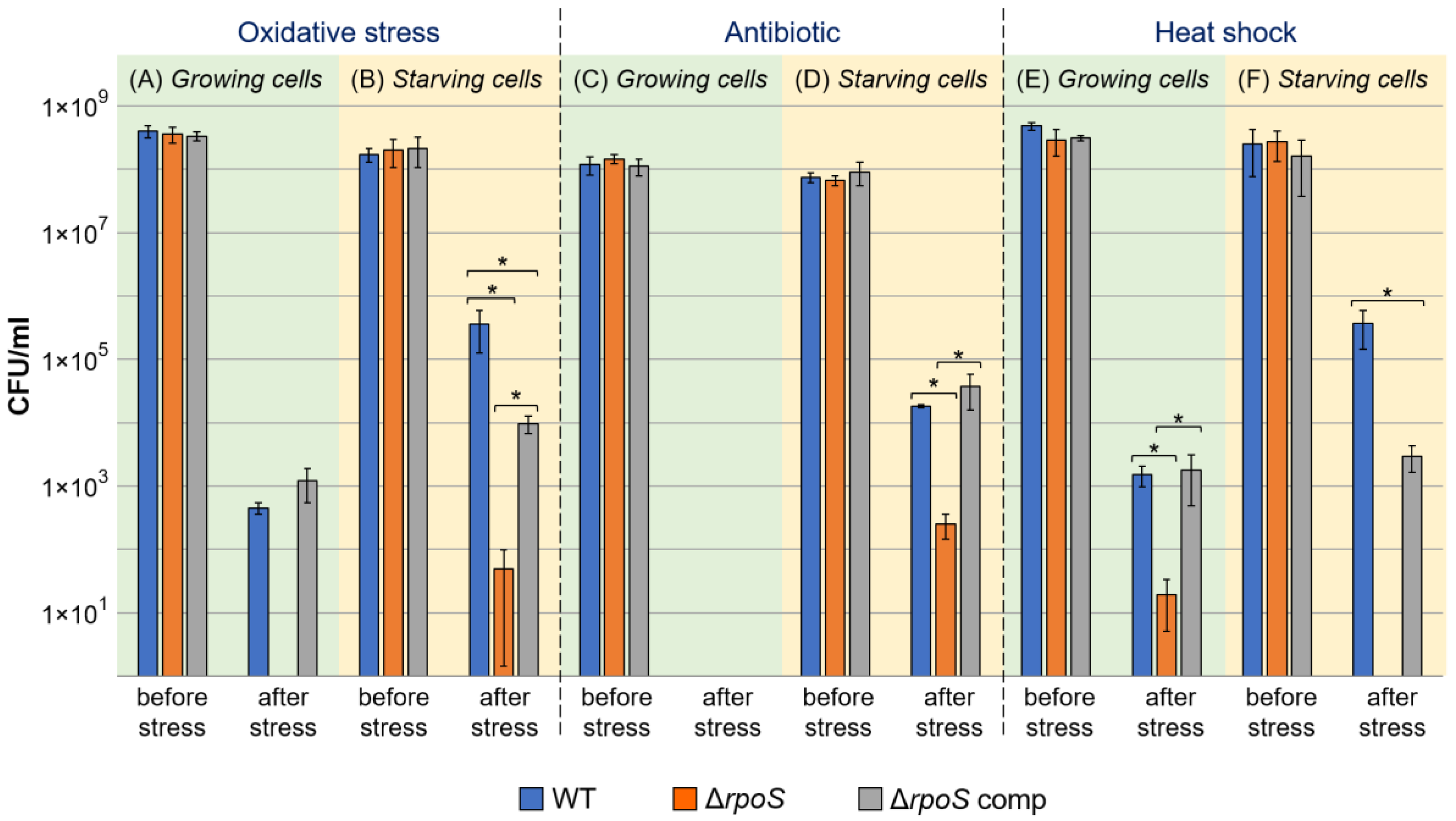
2.1.2. Cross-Protection
2.1.3. Virulence
2.2. Comparative Transcriptomics of WT and ΔrpoS Mutant under Growth and Starvation Conditions
2.2.1. Stress Genes
2.2.2. Transcription Factors
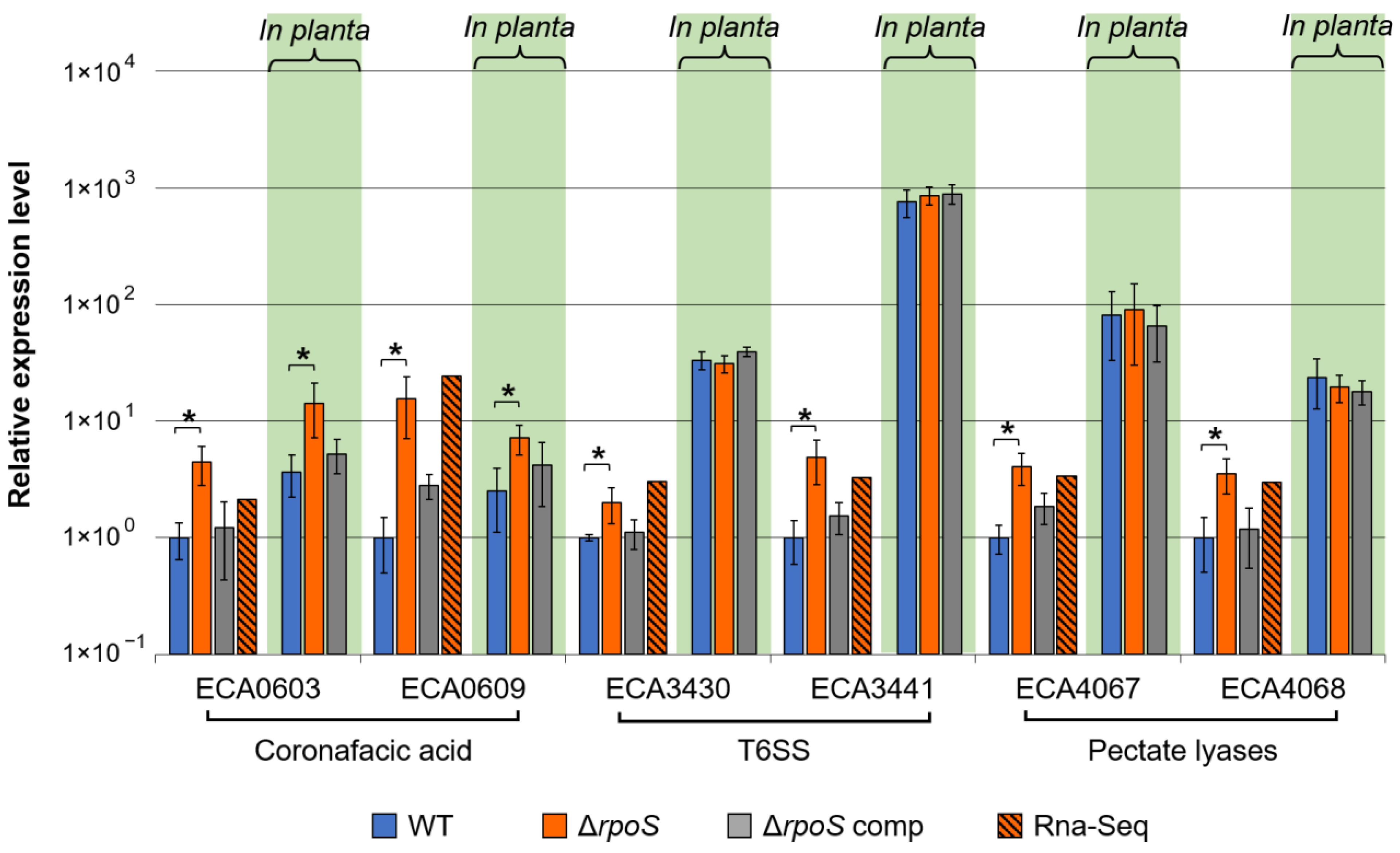
2.2.3. Virulence
2.2.4. General Metabolism and Transporters
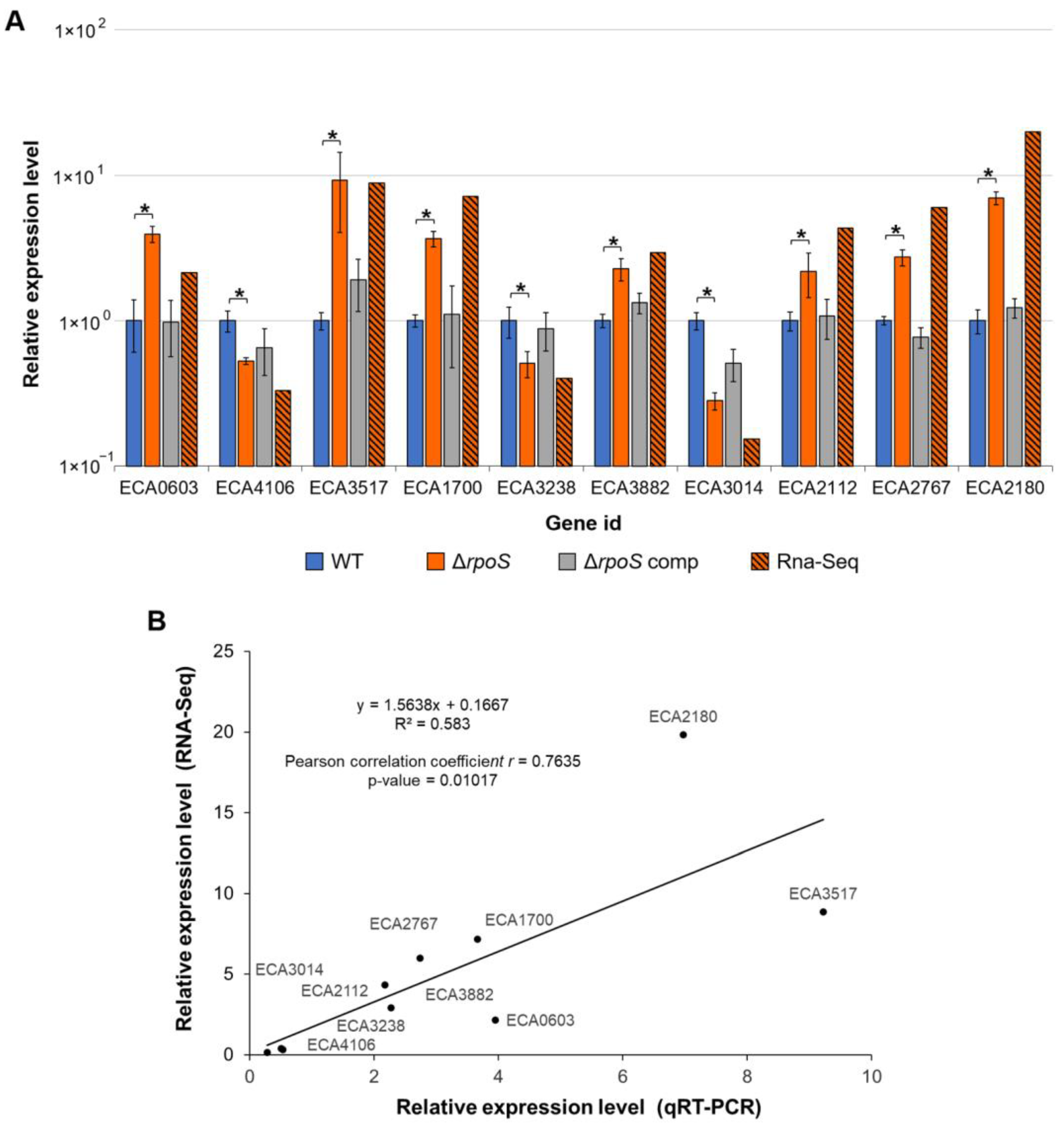
2.2.5. Verification of RNA-Seq Data
2.2.6. Genes of the P. atrosepticum RpoS Regulon
3. Materials and Methods
3.1. Bacterial Strains, Media, and Culture Conditions
3.2. Construction of rpoS Deletion Mutant
3.3. Construction of the ΔrpoS Complemented Mutant Strain Carrying the rpoS Gene on the Plasmid
3.4. Stress Tolerance Assay
3.5. Plant Cultivation and Infection
3.6. RNA Extraction
3.7. cDNA Library Preparation and Sequencing
3.8. qRT-PCR Analysis
3.9. Transcriptome Analysis
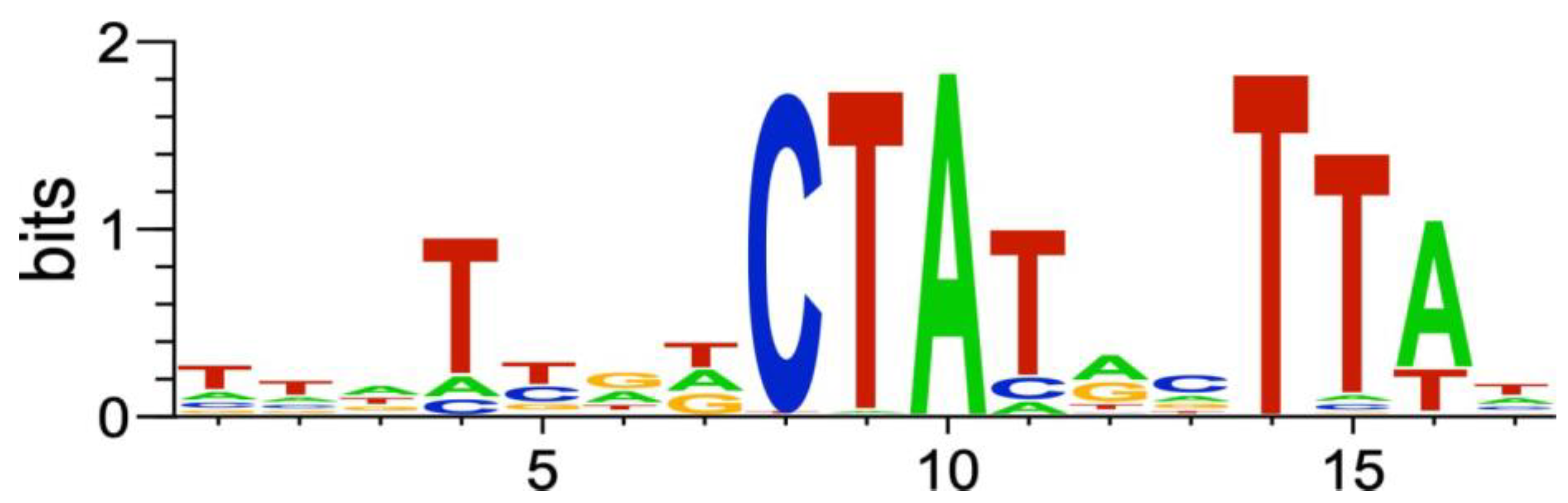
3.10. P. atrosepticum RpoS Regulon Inference
3.11. Motility Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Notley-McRobb, L.; King, T.; Ferenci, T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 2002, 184, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Chen, J.; Liu, R.; Zhang, X.H.; Jiang, Y.A. Mutation of rpoS gene decreased resistance to environmental stresses, synthesis of extracellular products and virulence of Vibrio anguillarum. FEMS Microbial. Ecol. 2009, 70, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.M.; Wang, W.; Silhavy, T.J. Novel RpoS-dependent mechanisms strengthen the envelope permeability barrier during stationary phase. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Velliou, E.G.; Van Derlinden, E.; Cappuyns, A.M.; Goossens, J.; Geeraerd, A.H.; Devlieghere, F.; Van Impe, J.F. Heat adaptation of Escherichia coli K12: Effect of acid and glucose. Procedia Food Sci. 2011, 1, 987–993. [Google Scholar] [CrossRef]
- Isohanni, P.; Huehn, S.; Aho, T.; Alter, T.; Lyhs, U. Heat stress adaptation induces cross-protection against lethal acid stress conditions in Arcobacter butzleri but not in Campylobacter jejuni. Food Microbiol. 2013, 34, 431–435. [Google Scholar] [CrossRef]
- Rosche, T.M.; Smith, D.J.; Parker, E.E.; Oliver, J.D. RpoS involvement and requirement for exogenous nutrient for osmotically induced cross protection in Vibrio vulnificus. FEMS Microbiol. Ecol. 2005, 53, 455–462. [Google Scholar] [CrossRef]
- Behmardi, P.; Grewal, E.; Kim, Y.; Yang, H. RpoS-dependant mechanism is required for cross-protection conferred to hyperosmolarity by heat shock. J. Exp. Microbiol. Immunol. 2009, 13, 18–21. [Google Scholar]
- Hagen, M.J.; Stockwell, V.O.; Whistler, C.A.; Johnson, K.B.; Loper, J.E. Stress tolerance and environmental fitness of Pseudomonas fluorescens A506, which has a mutation in RpoS. Phytopathology 2009, 99, 679–688. [Google Scholar] [CrossRef]
- Uhlich, G.A.; Chen, C.Y.; Cottrell, B.J.; Irwin, P.L.; Phillips, J.G. Peroxide resistance in Escherichia coli serotype O157: H7 biofilms is regulated by both RpoS-dependent and -independent mechanisms. Microbiology 2012, 158, 2225–2234. [Google Scholar] [CrossRef]
- Gayan, E.; Cambré, A.; Michiels, C.W.; Aertsen, A. RpoS-independent evolution reveals the importance of attenuated cAMP/CRP regulation in high hydrostatic pressure resistance acquisition in E. coli. Sci. Rep. 2017, 7, 8600. [Google Scholar] [CrossRef]
- Chiang, S.M.; Schellhorn, H.E. Evolution of the RpoS regulon: Origin of RpoS and the conservation of RpoS-dependent regulation in bacteria. J. Mol. Evol. 2010, 70, 557–571. [Google Scholar] [CrossRef] [PubMed]
- Santos-Zavaleta, A.; Gama-Castro, S.; Pérez-Rueda, E. A comparative genome analysis of the RpoS sigmulon shows a high diversity of responses and origins. Microbiology 2011, 157, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Franchini, A.G.; Ihssen, J.; Egli, T. Effect of global regulators RpoS and cyclic-AMP/CRP on the catabolome and transcriptome of Escherichia coli K12 during carbon-and energy-limited growth. PLoS ONE 2015, 10, e0133793. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, J.; Zhu, J.; Du, P.; Sun, A. Combined transcriptome and proteome analysis of RpoS regulon reveals its role in spoilage potential of Pseudomonas fluorescens. Front. Microbial. 2019, 10, 94. [Google Scholar] [CrossRef]
- Zhan, J.; Qiao, J.; Wang, X. Role of sigma factor RpoS in Cronobacter sakazakii environmental stress tolerance. Bioengineered 2021, 12, 2791–2809. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Schellhorn, H.E. Global effect of RpoS on gene expression in pathogenic Escherichia coli O157: H7 strain EDL933. BMC Genom. 2009, 10, 349. [Google Scholar] [CrossRef]
- Carter, M.Q.; Louie, J.W.; Huynh, S.; Parker, C.T. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157:H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol. 2014, 44, 108–118. [Google Scholar] [CrossRef]
- Dong, T.; Schellhorn, H.E. Role of RpoS in virulence of pathogens. Infect. Immun. 2010, 78, 887–897. [Google Scholar] [CrossRef]
- Flavier, A.B.; Schell, M.A.; Denny, T.P. An RpoS (σS) homologue regulates acylhomoserine lactone-dependent autoinduction in Ralstonia solanacearum. Mol. Microbial. 1998, 28, 475–486. [Google Scholar] [CrossRef]
- Mukherjee, A.; Cui, Y.; Ma, W.; Liu, Y.; Ishihama, A.; Eisenstark, A.; Chatterjee, A.K. RpoS (Sigma-S) controls expression of rsmA, a global regulator of secondary metabolites, harpin, and extracellular proteins in Erwinia carotovora. J. Bacteriol. 1998, 180, 3629–3634. [Google Scholar] [CrossRef]
- Andersson, R.A.; Kõiv, V.; Norman-Setterblad, C.; Pirhonen, M. Role of RpoS in virulence and stress tolerance of the plant pathogen Erwinia carotovora subsp. carotovora. Microbiology 1999, 145, 3547–3556. [Google Scholar] [CrossRef] [PubMed]
- Santander, R.D.; Monte-Serrano, M.; Rodríguez-Herva, J.J.; López-Solanilla, E.; Rodríguez-Palenzuela, P.; Biosca, E.G. Exploring new roles for the rpoS gene in the survival and virulence of the fire blight pathogen Erwinia amylovora. FEMS Microbiol. Ecol. 2014, 90, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Mattinen, L.; Tshuikina, M.; Mäe, A.; Pirhonen, M. Identification and characterization of Nip, necrosis-inducing virulence protein of Erwinia carotovora subsp. carotovora. Mol. Plant-Microbe Interact. 2004, 17, 1366–1375. [Google Scholar] [CrossRef]
- Li, Y.; Yamazaki, A.; Zou, L.; Biddle, E.; Zeng, Q.; Wang, Y.; Lin, H.; Wang, Q.; Yang, C.H. ClpXP protease regulates the type III secretion system of Dickeya dadantii 3937 and is essential for the bacterial virulence. Mol. Plant Microbe Interact. 2010, 23, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, A.; Li, J.; Hutchins, W.C.; Wang, L.; Ma, J.; Ibekwe, A.M.; Yang, C.H. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157: H7 EDL933 on lettuce leaves. Appl. Environ. Microbiol. 2011, 77, 156–162. [Google Scholar] [CrossRef]
- Lee, J.H.; Zhao, Y. ClpXP-dependent RpoS degradation enables full activation of type III secretion system, amylovoran production, and motility in Erwinia amylovora. Phytopathology 2017, 107, 1346–1352. [Google Scholar] [CrossRef]
- Fan, J.; Ma, L.; Zhao, C.; Yan, J.; Che, S.; Zhou, Z.; Wang, H.; Yang, L.; Hu, B. Transcriptome of Pectobacterium carotovorum subsp. carotovorum PccS1 infected in calla plants in vivo highlights a spatiotemporal expression pattern of genes related to virulence, adaptation, and host response. Mol. Plant Pathol. 2020, 21, 871–891. [Google Scholar] [CrossRef]
- Choi, O.; Kang, B.; Lee, Y.; Lee, Y.; Kim, J. Pantoea ananatis carotenoid production confers toxoflavin tolerance and is regulated by Hfq-controlled quorum sensing. Microbiol. Open 2021, 10, e1143. [Google Scholar] [CrossRef]
- Solis, R.; Bertani, I.; Degrassi, G.; Devescovi, G.; Venturi, V. Involvement of quorum sensing and RpoS in rice seedling blight caused by Burkholderia plantarii. FEMS Microbiol. Lett. 2006, 259, 106–112. [Google Scholar] [CrossRef]
- Gorshkov, V.; Petrova, O.; Gogoleva, N.; Gogolev, Y. Cell-to-cell communication in the populations of enterobacterium Erwinia carotovora ssp. atroseptica SCRI1043 during adaptation to stress conditions. FEMS Immunol. Med. Microbiol. 2010, 59, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.; Gorshkov, V.; Daminova, A.; Ageeva, M.; Moleleki, L.; Gogolev, Y. Stress response in Pectobacterium atrosepticum SCRI1043 under starvation conditions: Adaptive reactions at a low population density. Res. Microbiol. 2014, 165, 119–127. [Google Scholar] [CrossRef]
- Lange, R.; Hengge-Aronis, R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 1991, 5, 49–59. [Google Scholar] [CrossRef]
- Ramos-González, M.I.; Molin, S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J. Bacteriol. 1998, 180, 3421–3431. [Google Scholar] [CrossRef]
- O’Neal, C.R.; Gabriel, W.M.; Turk, A.K.; Libby, S.J.; Fang, F.C.; Spector, M.P. RpoS is necessary for both the positive and negative regulation of starvation survival genes during phosphate, carbon, and nitrogen starvation in Salmonella typhimurium. J. Bacteriol. 1994, 176, 4610–4616. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, A. Modulation of the nucleoid, the transcription apparatus, and the translation machinery in bacteria for stationary phase survival. Genes Cells 1999, 4, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, F.; Bally, M.; Chapon-Herve, V.; Michel, G.; Lazdunski, A.; Williams, P.; Stewart, G.S.A.B. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 1999, 145, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.J.; Silo-Suh, L.; Woods, D.E.; Hassett, D.J.; West, S.E.; Ohman, D.E. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 1999, 181, 3890–3897. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, V.; Kwenda, S.; Petrova, O.; Osipova, E.; Gogolev, Y.; Moleleki, L.N. Global gene expression analysis of cross-protected phenotype of Pectobacterium atrosepticum. PLoS ONE 2017, 12, e0169536. [Google Scholar] [CrossRef] [PubMed]
- Bearson, S.M.; Benjamin, W.H., Jr.; Swords, W.E.; Foster, J.W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J. Bacteriol. 1996, 178, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, V.; Parfirova, O.; Petrova, O.; Gogoleva, N.; Kovtunov, E.; Vorob’ev, V.; Gogolev, Y. The knockout of enterobactin-related gene in Pectobacterium atrosepticum results in reduced stress resistance and virulence towards the primed plants. Int. J. Mol. Sci. 2021, 22, 9594. [Google Scholar] [CrossRef]
- Tsers, I.; Parfirova, O.; Moruzhenkova, V.; Petrova, O.; Gogoleva, N.; Vorob’ev, V.; Gogolev, Y.; Gorshkov, V. A Switch from Latent to Typical Infection during Pectobacterium atrosepticum—Tobacco Interactions: Predicted and True Molecular Players. Int. J. Mol. Sci. 2023, 24, 13283. [Google Scholar] [CrossRef]
- Fang, F.C.; Libby, S.J.; Buchmeier, N.A.; Loewen, P.C.; Switala, J.; Harwood, J.; Guiney, D.G. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl. Acad. Sci. USA 1992, 89, 11978–11982. [Google Scholar] [CrossRef]
- Merrell, D.S.; Tischler, A.D.; Lee, S.H.; Camilli, A. Vibrio cholerae requires rpoS for efficient intestinal colonization. Infect. Immun. 2000, 68, 6691–6696. [Google Scholar] [CrossRef]
- Dong, T.; Coombes, B.K.; Schellhorn, H.E. Role of RpoS in the virulence of Citrobacter rodentium. Infect. Immun. 2009, 77, 501–507. [Google Scholar] [CrossRef]
- Mata, G.M.S.C.; Ferreira, G.M.; Spira, B. RpoS role in virulence and fitness in enteropathogenic Escherichia coli. PLoS ONE 2017, 12, e0180381. [Google Scholar] [CrossRef]
- Iriarte, M.; Stainier, I.; Cornelis, G.R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect. Immun. 1995, 63, 1840–1847. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Pan, Y.; Cai, Z.; Liu, Y.; Zhang, Y.; Liu, M.; Liu, Y.; Wang, K.; Zhang, L.; Yang, L. rpoS-mutation variants are selected in Pseudomonas aeruginosa biofilms under imipenem pressure. Cell Biosci. 2021, 11, 138. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Finkel, S.E. Dps protects cells against multiple stresses during stationary phase. J. Bacteriol. 2004, 186, 4192–4198. [Google Scholar] [CrossRef]
- Calhoun, L.N.; Kwon, Y.M. The effect of long-term propionate adaptation on the stress resistance of Salmonella Enteritidis. J. Appl. Microbiol. 2010, 109, 1294–1300. [Google Scholar] [CrossRef]
- Altuvia, S.; Almiron, M.; Huisman, G.; Kolter, R.; Storz, G. The dps promoter is activated by OxyR during growth and by IHF and σS in stationary phase. Mol. Microbiol. 1994, 13, 265–272. [Google Scholar] [CrossRef]
- Zafar, M.A.; Carabetta, V.J.; Mandel, M.J.; Silhavy, T.J. Transcriptional occlusion caused by overlapping promoters. Proc. Nat. Acad. Sci. 2014, 111, 1557–1561. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, G.P.; Creighton, R.I.; Nikolaev, Y.; Booth, I.R. Importance of RpoS and Dps in survival of exposure of both exponential-and stationary-phase Escherichia coli cells to the electrophile N-ethylmaleimide. J. Bacteriol. 1998, 180, 1030–1036. [Google Scholar] [CrossRef] [PubMed]
- White-Ziegler, C.A.; Um, S.; Perez, N.M.; Berns, A.L.; Malhowski, A.J.; Young, S. Low temperature (23 degrees C) increases expression of biofilm-, cold-shock-and RpoS-dependent genes in Escherichia coli K-12. Microbiology 2008, 154, 148. [Google Scholar] [CrossRef]
- Christensen, S.K.; Maenhaut-Michel, G.; Mine, N.; Gottesman, S.; Gerdes, K.; Van Melderen, L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 2004, 51, 1705–1717. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; Cherny, I.; Khoo, S.K.; de Lacoba, M.G.; Chan, W.T.; Yeo, C.C.; Gazit, E.; Espinosa, M. The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: Functional and structural correlation. J. Bacteriol. 2007, 186, 1266–1278. [Google Scholar] [CrossRef]
- Cho, S.H.; Szewczyk, J.; Pesavento, C.; Zietek, M.; Banzhaf, M.; Roszczenko, P.; Asmar, A.; Laloux, G.; Hov, A.K.; Leverrier, P.; et al. Detecting envelope stress by monitoring β-barrel assembly. Cell 2014, 159, 1652–1664. [Google Scholar] [CrossRef]
- Wall, E.A.; Majdalani, N.; Gottesman, S. IgaA negatively regulates the Rcs phosphorelay via contact with the RcsD phosphotransfer protein. PLoS Genet. 2020, 16, e1008610. [Google Scholar] [CrossRef]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010, 391, 209–213. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, X.; Zhang, X.S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef]
- Schmid, B.; Klumpp, J.; Raimann, E.; Loessner, M.J.; Stephan, R.; Tasara, T. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 2009, 75, 1621–1627. [Google Scholar] [CrossRef]
- Loepfe, C.; Raimann, E.; Stephan, R.; Tasara, T. Reduced host cell invasiveness and oxidative stress tolerance in double and triple csp gene family deletion mutants of Listeria monocytogenes. Foodborne Pathog. Dis. 2010, 7, 775–783. [Google Scholar] [CrossRef]
- Piróg, A.; Cantini, F.; Nierzwicki, Ł.; Obuchowski, I.; Tomiczek, B.; Czub, J.; Liberek, K. Two bacterial small heat shock proteins, IbpA and IbpB, form a functional heterodimer. J. Mol. Biol. 2021, 433, 167054. [Google Scholar] [CrossRef] [PubMed]
- Fridovich, I. Superoxide radical: An endogenous toxicant. Annu. Rev. Pharmacol. Toxicol. 1983, 23, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Gort, A.S.; Ferber, D.M.; Imlay, J.A. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol. Microbiol. 1999, 32, 179–191. [Google Scholar] [CrossRef]
- Kong, L.; Xiong, Z.; Song, X.; Xia, Y.; Zhang, H.; Yang, Y.; Ai, L. Enhanced antioxidant activity in Streptococcus thermophilus by high-level expression of superoxide dismutase. Front. Microbiol. 2020, 11, 579804. [Google Scholar] [CrossRef] [PubMed]
- Farewell, A.; Kvint, K.; Nyström, T. uspB, a new ςS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J. Bacteriol. 1998, 180, 6140–6147. [Google Scholar] [CrossRef] [PubMed]
- Tramonti, A.; Visca, P.; De Canio, M.; Falconi, M.; De Biase, D. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 2002, 184, 2603–2613. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Schellhorn, H.E. Controlled induction of the RpoS regulon in Escherichia coli, using an RpoS-expressing plasmid. Can. J. Microbiol. 2003, 49, 733–740. [Google Scholar] [CrossRef]
- Ma, Z.; Richard, H.; Foster, J.W. pH-Dependent modulation of cyclic AMP levels and GadW-dependent repression of RpoS affect synthesis of the GadX regulator and Escherichia coli acid resistance. J. Bacteriol. 2003, 185, 6852–6859. [Google Scholar] [CrossRef]
- Abdallah, F.B.; Chaieb, K.; Snoussi, M.; Bakhrouf, A.; Gaddour, K. Phenotypic variations and molecular identification of Salmonella enterica serovar Typhimurium cells under starvation in seawater. Curr. Microbiol. 2007, 55, 485–491. [Google Scholar] [CrossRef]
- Cho, B.K.; Kim, D.; Knight, E.M.; Zengler, K.; Palsson, B.O. Genome-scale reconstruction of the sigma factor network in Escherichia coli: Topology and functional states. BMC Biol. 2014, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Marschall, C.; Labrousse, V.; Kreimer, M.; Weichart, D.; Kolb, A.; Hengge-Aronis, R. Molecular analysis of the regulation of csiD, a carbon starvation-inducible gene in Escherichia coli that is exclusively dependent on σS and requires activation by cAMP-CRP. J. Mol. Biol. 1998, 276, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Colland, F.; Barth, M.; Hengge-Aronis, R.; Kolb, A. σ factor selectivity of Escherichia coli RNA polymerase: Role for CRP, IHF and Lrp transcription factors. EMBO J. 2000, 19, 3028–3037. [Google Scholar] [CrossRef]
- Weber, H.; Polen, T.; Heuveling, J.; Wendisch, V.F.; Hengge, R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005, 187, 1591–1603. [Google Scholar] [CrossRef]
- Mason, C.; Thompson, C.; Ouyang, Z. DksA plays an essential role in regulating the virulence of Borrelia burgdorferi. Mol. Microbiol. 2020, 114, 172–183. [Google Scholar] [CrossRef]
- Boyle, W.K.; Richards, C.L.; Dulebohn, D.P.; Zalud, A.K.; Shaw, J.A.; Lovas, S.; Gherardini, F.C.; Bourret, T.J. DksA-dependent regulation of RpoS contributes to Borrelia burgdorferi tick-borne transmission and mammalian infectivity. PLoS Pathog. 2021, 17, e1009072. [Google Scholar] [CrossRef]
- Kravchenko, U.; Gogoleva, N.; Kalubaka, N.; Kruk, A.; Diubo, Y.; Gogolev, Y.; Nikolaichik, Y. The PhoPQ two-component system is the major regulator of cell surface properties, stress responses and plant-derived substrate utilisation during development of pectobacterium versatile-host plant pathosystems. Front. Microbiol. 2021, 11, 621391. [Google Scholar] [CrossRef]
- Mattinen, L.; Nissinen, R.; Riipi, T.; Kalkkinen, N.; Pirhonen, M. Host-extract induced changes in the secretome of the plant pathogenic bacterium Pectobacterium atrosepticum. Proteomics 2007, 7, 3527–3537. [Google Scholar] [CrossRef]
- Liu, H.; Coulthurst, S.J.; Pritchard, L.; Hedley, P.E.; Ravensdale, M.; Humphris, S.; Burr, T.; Takle, G.; Brurberg, M.B.; Birch, P.R.J.; et al. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 2008, 4, e1000093. [Google Scholar] [CrossRef]
- Weber, B.; Hasic, M.; Chen, C.; Wai, S.N.; Milton, D.L. Type VI secretion modulates quorum sensing and stress response in Vibrio anguillarum. Environ. Microbiol. 2009, 11, 3018–3028. [Google Scholar] [CrossRef]
- Gueguen, E.; Durand, E.; Zhang, X.Y.; d’Amalric, Q.; Journet, L.; Cascales, E. Expression of a Yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS ONE 2013, 8, e66615. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Song, Y.; Wang, T.; Xu, S.; Peng, Z.; Lin, X.; Zhang, L.; Shen, X. A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ. Microbiol. 2013, 15, 557–569. [Google Scholar] [CrossRef]
- Guan, J.; Xiao, X.; Xu, S.; Gao, F.; Wang, J.; Wang, T.; Song, Y.; Pan, J.; Shen, X.; Wang, Y. Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J. Microbiol. 2015, 53, 633–642. [Google Scholar] [CrossRef]
- Gorshkov, V.; Gubaev, R.; Petrova, O.; Daminova, A.; Gogoleva, N.; Ageeva, M.; Parfirova, O.; Prokchorchik, M.; Nikolaichik, Y.; Gogolev, Y. Transcriptome profiling helps to identify potential and true molecular switches of stealth to brute force behavior in Pectobacterium atrosepticum during systemic colonization of tobacco plants. Eur. J. Plant Pathol. 2018, 152, 957–976. [Google Scholar] [CrossRef]
- Gorshkov, V.Y.; Toporkova, Y.Y.; Tsers, I.D.; Smirnova, E.O.; Ogorodnikova, A.V.; Gogoleva, N.E.; Parfirova, O.I.; Petrova, O.E.; Gogolev, Y.V. Differential modulation of the lipoxygenase cascade during typical and latent Pectobacterium atrosepticum infections. Ann. Bot. 2022, 129, 271–286. [Google Scholar] [CrossRef]
- Nielsen, A.T.; Dolganov, N.A.; Otto, G.; Miller, M.C.; Wu, C.Y.; Schoolnik, G.K. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2006, 2, e109. [Google Scholar] [CrossRef]
- Dong, T.; Yu, R.; Schellhorn, H. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 2011, 79, 375–386. [Google Scholar] [CrossRef]
- Dudin, O.; Geiselmann, J.; Ogasawara, H.; Ishihama, A.; Lacour, S. Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J. Bacteriol. 2014, 196, 707–715. [Google Scholar] [CrossRef]
- Makinoshima, H.; Aizawa, S.I.; Hayashi, H.; Miki, T.; Nishimura, A.; Ishihama, A. Growth phase-coupled alterations in cell structure and function of Escherichia coli. J. Bacteriol. 2003, 185, 1338–1345. [Google Scholar] [CrossRef]
- Klotz, A.; Forchhammer, K. Glycogen, a major player for bacterial survival and awakening from dormancy. Future Microbiol. 2017, 12, 101–104. [Google Scholar] [CrossRef]
- Metaane, S.; Monteil, V.; Ayrault, S.; Bordier, L.; Levi-Meyreuis, C.; Norel, F. The stress sigma factor σS/RpoS counteracts Fur repression of genes involved in iron and manganese metabolism and modulates the ionome of Salmonella enterica serovar Typhimurium. PLoS ONE 2022, 17, e0265511. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, G.; Cui, Y.; Mukherjee, A.; Ma, W.L.; Chatterjee, A.K. kdgREcc Negatively Regulates Genes for Pectinases, Cellulase, Protease, HarpinEcc, and a Global RNA Regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 1999, 181, 2411–2422. [Google Scholar] [CrossRef]
- Tarasova, N.; Gorshkov, V.; Petrova, O.; Gogolev, Y. Potato signal molecules that activate pectate lyase synthesis in Pectobacterium atrosepticum SCRI1043. World J. Microbiol. Biotechnol. 2013, 29, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Nikolaichik, Y.; Damienikan, A.U. SigmoID: A user-friendly tool for improving bacterial genome annotation through analysis of transcription control signals. PeerJ 2016, 4, e2056. [Google Scholar] [CrossRef]
- Wong, G.T.; Bonocora, R.P.; Schep, A.N.; Beeler, S.M.; Lee Fong, A.J.; Shull, L.M.; Batachari, L.E.; Dillon, M.; Evans, C.; Becker, C.J.; et al. Genome-wide transcriptional response to varying RpoS levels in Escherichia coli K-12. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Tierrafría, V.H.; Rioualen, C.; Salgado, H.; Lara, P.; Gama-Castro, S.; Lally, P.; Gómez-Romero, L.; Peña-Loredo, P.; López-Almazo, A.G.; Alarcón-Carranza, G.; et al. RegulonDB 11.0: Comprehensive high-throughput datasets on transcriptional regulation in Escherichia coli K-12. Microb. Genom. 2022, 8, mgen000833. [Google Scholar] [CrossRef] [PubMed]
- Kaniga, K.; Delor, I.; Cornelis, G.R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: Inactivation of the blaA gene of Yersinia enterocolitica. Gene 1991, 109, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Quan, J.; Tian, J. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 2009, 4, e6441. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, Menlo Park, CA, USA, 31 December 1994; pp. 28–36. [Google Scholar]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Tomar, T.S.; Dasgupta, P.; Kanaujia, S.P. Operon Finder: A Deep Learning-based Web Server for Accurate Prediction of Prokaryotic Operons. J. Mol. Biol. 2023, 435, 167921. [Google Scholar] [CrossRef]
- Persson, E.; Sonnhammer, E.L. InParanoid-DIAMOND: Faster orthology analysis with the InParanoid algorithm. Bioinformatics 2022, 38, 2918–2919. [Google Scholar] [CrossRef]
- Chang, L.; Wei, L.I.C.; Audia, J.P.; Morton, R.A.; Schellhorn, H.E. Expression of the Escherichia coli NRZ nitrate reductase is highly growth phase dependent and is controlled by RpoS, the alternative vegetative sigma factor. Mol. Microbiol. 1999, 34, 756–766. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Audia, J.P.; Roy, R.N.; Schellhorn, H.E. Transcriptional induction of the conserved alternative sigma factor RpoS in Escherichia coli is dependent on BarA, a probable two-component regulator. Mol. Microbiol. 2000, 37, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.S.; Sebaihia, M.; Pritchard, L.; Holden, M.T.G.; Hyman, L.J.; Holeva, M.C.; Thomson, N.R.; Bentley, S.D.; Churcher, L.J.C.; Mungall, K.; et al. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 2004, 101, 11105–11110. [Google Scholar] [CrossRef] [PubMed]
- Herrero, M.; de Lorenzo, V.; Timmis, K.N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 1990, 172, 6557–6567. [Google Scholar] [CrossRef] [PubMed]
- Grinter, N.J. A broad-host-range cloning vector transposable to various replicons. Gene 1983, 21, 133–143. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]


| Variant of Comparison | Total DEGs | Upregulated DEGs | Downregulated DEGs |
|---|---|---|---|
| WT_S/WT_G | 2222 | 1075 | 1147 |
| Mu_S/Mu_G | 1934 | 968 | 966 |
| Mu_S/WT_S | 615 | 389 | 226 |
| Mu_G/WT_G | 359 | 200 | 159 |
| Expression Level, log2FC | ||||||
|---|---|---|---|---|---|---|
| Locus Tag | Gene | Annotation | WT_S/ WT_G | Mu_S/ Mu_G | Mu_S/ WT_S | Mu_G/ WT_G |
| ECA2767 | dps | DNA starvation/stationary phase protection protein Dps | nD | 3.31 | 2.58 | nD |
| ECA0443 | Txe/YoeB family addiction module toxin | 1.29 | 1.71 | 1.11 | nD | |
| ECA0900 | tehB | SAM-dependent methyltransferase TehB | nD | 2.69 | 1.83 | nD |
| ECA3053 | cvpA | colicin V production protein | nD | 1.55 | 2.11 | nD |
| ECA1580 | cold-shock domain-containing protein | −1.03 | nD | 1.1 | nD | |
| ECA3527 | rcsF | Rcs stress response system protein RcsF | −3.63 | −1.82 | 1.48 | nD |
| ECA2659 | cspD | cold-shock-like protein CspD | nD | nD | 1.54 | 1.46 |
| ECA4402 | ibpB | heat-shock chaperone IbpB | nD | 2.89 | 2.45 | −1.07 |
| ECA4403 | ibpA | heat-shock chaperone IbpA | nD | 3.34 | 2.75 | nD |
| Expression Level, log2FC | ||||||
|---|---|---|---|---|---|---|
| Locus Tag | Gene | Annotation | WT_S/ WT_G | Mu_S/ Mu_G | Mu_S/ WT_S | Mu_G/ WT_G |
| ECA3410 | type I toxin–antitoxin system SymE family toxin | 1.09 | nD | −6.39 | −6.72 | |
| ECA2211 | sodC | copper-zinc superoxide dismutase | nD | nD | −2.19 | −2.16 |
| ECA0049 | uspB | universal stress protein UspB | nD | nD | −1.66 | −1.62 |
| ECA3409 | type I toxin–antitoxin system SymE family toxin | 1.03 | 1.06 | 1.01 | 1.08 | |
| ECA2659 | cspD | cold-shock-like protein CspD | nD | nD | 1.54 | 1.46 |
| Expression Level, log2FC | ||||||
|---|---|---|---|---|---|---|
| Locus Tag | Gene | Annotation | WT_S/ WT_G | Mu_S/ Mu_G | Mu_S/ WT_S | Mu_G/ WT_G |
| ECA2724 | rscR | LysR-family transcriptional regulator | 1.28 | nD | −1.95 | nD |
| ECA0739 | budR | LysR-family transcriptional regulator | 1.33 | nD | −1.03 | nD |
| ECA1806 | helix-turn-helix domain-containing protein | 1.42 | nD | −1.24 | nD | |
| ECA0643 | exuR | exu regulon transcriptional regulator | 1.42 | nD | −1.21 | nD |
| ECA1985 | pspC | phage shock protein C | 1.47 | nD | −1.16 | nD |
| ECA0204 | rfaH | transcription/translation regulatory transformer protein RfaH | 1.58 | nD | −1.23 | nD |
| ECA0582 | type II toxin–antitoxin system PemK/MazF family toxin | 1.61 | nD | −1.04 | nD | |
| ECA4389 | PRD domain-containing protein | 1.63 | nD | −1.15 | nD | |
| ECA1849 | GntR-family transcriptional regulator | 1.64 | nD | −1.00 | nD | |
| ECA0008 | qseB | two-component system response regulator | 1.66 | nD | −1.40 | nD |
| ECA1194 | cueR | copper efflux regulator | 1.70 | nD | −1.64 | nD |
| ECA3469 | XRE family transcriptional regulator | 2.06 | nD | −1.08 | nD | |
| ECA4123 | rexZ | regulator of exoenzymes | 4.36 | 3.72 | −1.02 | nD |
| ECA0673 | XRE family transcriptional regulator | 4.43 | 3.30 | −1.02 | nD | |
| ECA3238 | iscR | Fe-S cluster assembly transcriptional regulator IscR | 4.51 | 3.80 | −1.32 | nD |
| ECA0064 | type II toxin–antitoxin system Phd/YefM family antitoxin | 4.56 | 2.84 | −1.75 | nD | |
| ECA4381 | lgoR | L-galactonate utilization transcriptional regulator | 5.47 | 3.68 | −1.90 | nD |
| Expression Level, log2FC | ||||||
|---|---|---|---|---|---|---|
| Locus Tag | Gene | Annotation | WT_S/ WT_G | Mu_S/ Mu_G | Mu_S/ WT_S | Mu_G/ WT_G |
| ECA4154 | putative transcriptional regulator | 1.03 | 2.68 | 1.60 | nD | |
| ECA3959 | ArsR-family transcriptional regulator | 1.07 | 1.86 | 1.43 | nD | |
| ECA0180 | metR | transcriptional activator protein | 1.54 | 4.43 | 3.12 | nD |
| ECA2069 | TetR-family transcriptional regulator | 1.95 | 2.60 | 1.25 | nD | |
| ECA2295 | AraC-family transcriptional regulator | 1.95 | 3.52 | 1.68 | nD | |
| ECA0903 | norR | nitric oxide reductase sigma-54-dependent transcriptional regulator | 2.22 | 3.09 | 1.18 | nD |
| ECA2910 | MarR family transcriptional regulator | 3.49 | 5.73 | 2.11 | nD | |
| ECA2873 | putative transcriptional regulator | 3.82 | 6.42 | 2.70 | nD | |
| Gene | Locus Tag | Product | Functional Supercategory | |
|---|---|---|---|---|
| P. atrosepticum | E. coli | |||
| osmY | ECA0469 | b4376 | periplasmic chaperone | Stress |
| alkB | ECA0909 | b2212 | DNA oxidative demethylase | General metabolism |
| galE | ECA1389 | b0759 | UDP-glucose 4-epimerase | General metabolism |
| galM | ECA1388 | b0756 | galactose-1-epimerase | General metabolism |
| pykF | ECA1867 | b1676 | pyruvate kinase | General metabolism |
| sodC | ECA2211 | b1646 | superoxide dismutase (Cu-Zn) | Stress |
| adhE | ECA2326 | b1241 | fused acetaldehyde-CoA dehydrogenase and iron-dependent alcohol dehydrogenasealdehyde/alcohol dehydrogenase | General metabolism, stress |
| elaB | ECA3015 | b2266 | tail-anchored inner membrane protein | Stress |
| frdA | ECA3969 | b4154 | fumarate reductase flavoprotein subunit | General metabolism |
| frdB | ECA3970 | b4153 | fumarate reductase iron-sulfur protein | General metabolism |
| frdC | ECA3971 | b4152 | fumarate reductase membrane protein FrdC | General metabolism |
| frdD | ECA3972 | b4151 | fumarate reductase membrane protein FrdD | General metabolism |
| ecnB | ECA3975 | b4411 | bacteriolytic entericidin B lipoprotein | Stress |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrova, O.; Semenova, E.; Parfirova, O.; Tsers, I.; Gogoleva, N.; Gogolev, Y.; Nikolaichik, Y.; Gorshkov, V. RpoS-Regulated Genes and Phenotypes in the Phytopathogenic Bacterium Pectobacterium atrosepticum. Int. J. Mol. Sci. 2023, 24, 17348. https://doi.org/10.3390/ijms242417348
Petrova O, Semenova E, Parfirova O, Tsers I, Gogoleva N, Gogolev Y, Nikolaichik Y, Gorshkov V. RpoS-Regulated Genes and Phenotypes in the Phytopathogenic Bacterium Pectobacterium atrosepticum. International Journal of Molecular Sciences. 2023; 24(24):17348. https://doi.org/10.3390/ijms242417348
Chicago/Turabian StylePetrova, Olga, Elizaveta Semenova, Olga Parfirova, Ivan Tsers, Natalia Gogoleva, Yuri Gogolev, Yevgeny Nikolaichik, and Vladimir Gorshkov. 2023. "RpoS-Regulated Genes and Phenotypes in the Phytopathogenic Bacterium Pectobacterium atrosepticum" International Journal of Molecular Sciences 24, no. 24: 17348. https://doi.org/10.3390/ijms242417348








