Differential Expression of LLT1, SLAM Receptors CS1 and 2B4 and NCR Receptors NKp46 and NKp30 in Pediatric Acute Lymphoblastic Leukemia (ALL)
Abstract
:1. Introduction
2. Results
2.1. RNA Seq Expression Analysis of Immune Receptors in B-ALL and T-ALL Subjects
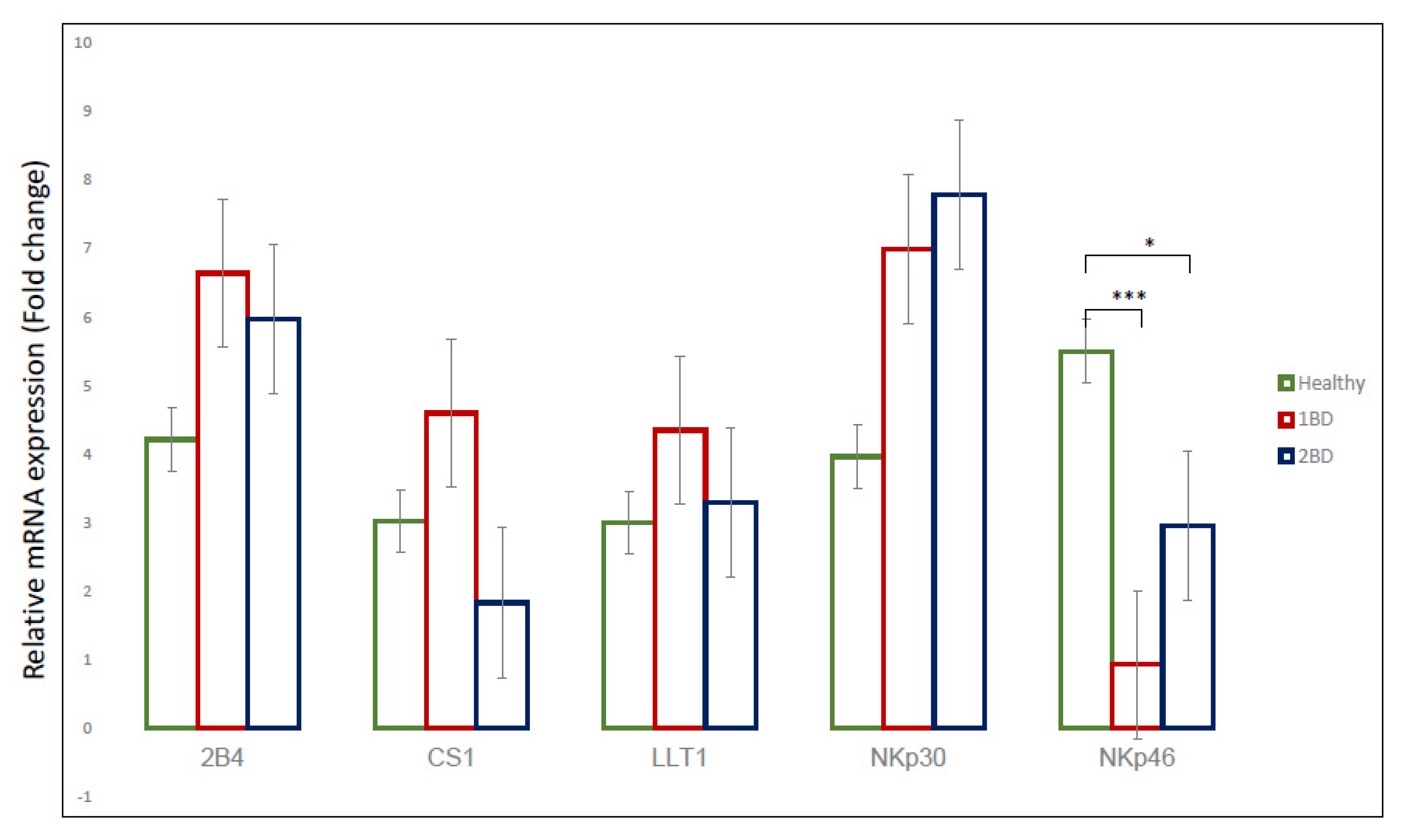
2.2. mRNA Expression of Immune Receptors in Pediatric ALL Subjects
2.3. Differential Cell Surface Expression of Immune Receptors on Monocytes (CD14+ Cells) in ALL Subjects
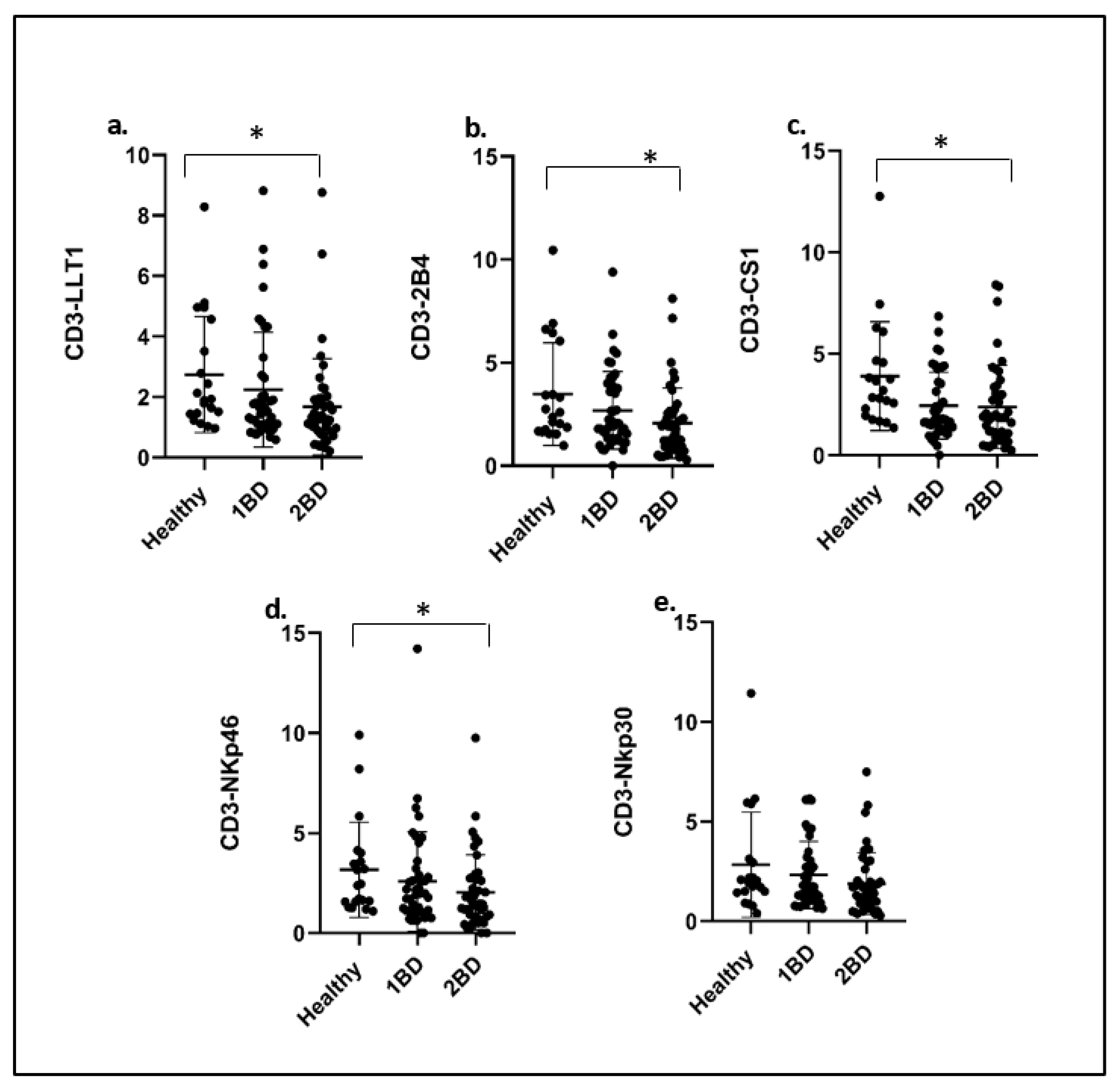
2.4. Downregulation of Cell Surface Expression of Immune Receptors on T Cells (CD3+) of ALL Subjects after Treatment
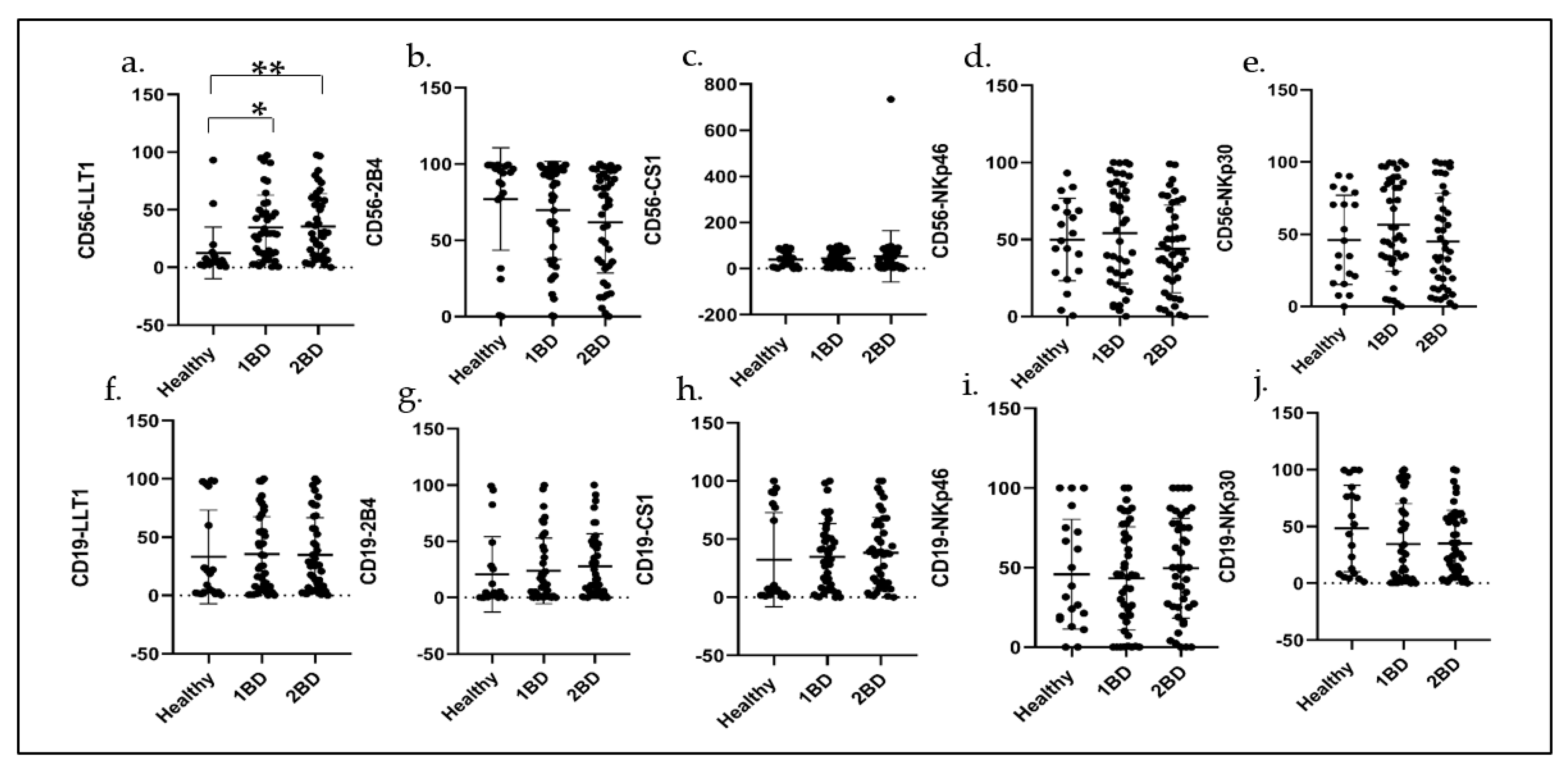
2.5. Cell Surface Expression of Immune Receptors in CD56+ NK Cells and CD19+ B Cells in ALL Subjects before and after Induction Chemotherapy
2.6. Demographic Data of ALL Subjects
3. Discussion
4. Materials and Methods
4.1. RNA Sequencing Data Collection
4.2. Subjects and Healthy Volunteers
4.3. Blood Collection
4.4. PBMC Isolation
4.5. qRT-PCR Analysis
4.6. Flow Cytometry
4.7. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Advani, A. Acute lymphoblastic leukemia (ALL). Best Pract. Res. Clin. Haematol. 2017, 30, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Aldoss, I.; Forman, S.J.; Pullarkat, V. Acute Lymphoblastic Leukemia in the Older Adult. J. Oncol. Pract. 2019, 15, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Fielding, A.K.; Richards, S.M.; Chopra, R.; Lazarus, H.M.; Litzow, M.R.; Buck, G.; Durrant, I.J.; Luger, S.M.; Marks, D.I.; Franklin, I.M.; et al. Outcome of 609 adults after relapse of acute lymphoblastic leukemia (ALL); an MRC UKALL12/ECOG 2993 study. Blood 2007, 109, 944–950. [Google Scholar] [CrossRef] [Green Version]
- Larson, R.A. Managing CNS disease in adults with acute lymphoblastic leukemia. Leuk. Lymphoma 2018, 59, 3–13. [Google Scholar] [CrossRef]
- Stelljes, M.; Marks, D.I. Acute Lymphoblastic Leukemia in Adults. In The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies, 7th ed.; Carreras, E., Dufour, C., Mohty, M., Kroger, N., Eds.; Springer: Cham, Switzerland, 2019; pp. 531–538. [Google Scholar]
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [Green Version]
- Inaba, H.; Greaves, M.; Mullighan, C.G. Acute lymphoblastic leukaemia. Lancet 2013, 381, 1943–1955. [Google Scholar] [CrossRef] [Green Version]
- Pui, C.H.; Yang, J.J.; Hunger, S.P.; Pieters, R.; Schrappe, M.; Biondi, A.; Vora, A.; Baruchel, A.; Silverman, L.B.; Schmiegelow, K.; et al. Childhood Acute Lymphoblastic Leukemia: Progress Through Collaboration. J. Clin. Oncol. 2015, 33, 2938–2948. [Google Scholar] [CrossRef] [Green Version]
- Maloney, K.W.; Gore, L. Agents in Development for Childhood Acute Lymphoblastic Leukemia. Paediatr. Drugs 2018, 20, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Velentza, L.; Zaman, F.; Savendahl, L. Bone health in glucocorticoid-treated childhood acute lymphoblastic leukemia. Crit. Rev. Oncol. Hematol. 2021, 168, 103492. [Google Scholar] [CrossRef]
- Stanulla, M.; Schrappe, M. Treatment of childhood acute lymphoblastic leukemia. Semin. Hematol. 2009, 46, 52–63. [Google Scholar] [CrossRef]
- Inaba, H.; Mullighan, C.G. Pediatric acute lymphoblastic leukemia. Haematologica 2020, 105, 2524–2539. [Google Scholar] [CrossRef] [PubMed]
- Berkman, A.M.; Andersen, C.R.; Cuglievan, B.; McCall, D.C.; Lupo, P.J.; Parsons, S.K.; DiNardo, C.D.; Short, N.J.; Jain, N.; Kadia, T.M.; et al. Long-Term Outcomes among Adolescent and Young Adult Survivors of Acute Leukemia: A Surveillance, Epidemiology, and End Results Analysis. Cancer Epidemiol. Biomark. Prev. 2022, 31, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.P.; Raetz, E.A. How I treat relapsed acute lymphoblastic leukemia in the pediatric population. Blood 2020, 136, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Carroll, W.L.; Hunger, S.P. Therapies on the horizon for childhood acute lymphoblastic leukemia. Curr. Opin. Pediatr. 2016, 28, 12–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeAngelo, D.J.; Jabbour, E.; Advani, A. Recent Advances in Managing Acute Lymphoblastic Leukemia. Am. Soc. Clin. Oncol. Educ. Book 2020, 40, 330–342. [Google Scholar] [CrossRef]
- DasGupta, R.K.; Marini, B.L.; Rudoni, J.; Perissinotti, A.J. A review of CD19-targeted immunotherapies for relapsed or refractory acute lymphoblastic leukemia. J. Oncol. Pharm. Pract. 2018, 24, 453–467. [Google Scholar] [CrossRef]
- Kimble, E.L.; Cassaday, R.D. Antibody and cellular immunotherapies for acute lymphoblastic leukemia in adults. Leuk. Lymphoma 2021, 62, 3333–3347. [Google Scholar] [CrossRef]
- Romanski, A.; Bug, G.; Becker, S.; Kampfmann, M.; Seifried, E.; Hoelzer, D.; Ottmann, O.G.; Tonn, T. Mechanisms of resistance to natural killer cell-mediated cytotoxicity in acute lymphoblastic leukemia. Exp. Hematol. 2005, 33, 344–352. [Google Scholar] [CrossRef]
- Liu, S.; Galat, V.; Galat, Y.; Lee, Y.K.A.; Wainwright, D.; Wu, J. NK cell-based cancer immunotherapy: From basic biology to clinical development. J. Hematol. Oncol. 2021, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Carlsten, M.; Jaras, M. Natural Killer Cells in Myeloid Malignancies: Immune Surveillance, NK Cell Dysfunction, and Pharmacological Opportunities to Bolster the Endogenous NK Cells. Front. Immunol. 2019, 10, 2357. [Google Scholar] [CrossRef]
- Oelsner, S.; Waldmann, A.; Billmeier, A.; Roder, J.; Lindner, A.; Ullrich, E.; Marschalek, R.; Dotti, G.; Jung, G.; Grosse-Hovest, L.; et al. Genetically engineered CAR NK cells display selective cytotoxicity against FLT3-positive B-ALL and inhibit in vivo leukemia growth. Int. J. Cancer 2019, 145, 1935–1945. [Google Scholar] [CrossRef] [PubMed]
- Paust, S.; Blish, C.A.; Reeves, R.K. Redefining Memory: Building the Case for Adaptive NK Cells. J. Virol. 2017, 91, e00169-17. [Google Scholar] [CrossRef] [Green Version]
- Pahl, J.H.W.; Koch, J.; Gotz, J.J.; Arnold, A.; Reusch, U.; Gantke, T.; Rajkovic, E.; Treder, M.; Cerwenka, A. CD16A Activation of NK Cells Promotes NK Cell Proliferation and Memory-Like Cytotoxicity against Cancer Cells. Cancer Immunol. Res. 2018, 6, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Becker-Hapak, M.K.; Shrestha, N.; McClain, E.; Dee, M.J.; Chaturvedi, P.; Leclerc, G.M.; Marsala, L.I.; Foster, M.; Schappe, T.; Tran, J.; et al. A Fusion Protein Complex that Combines IL-12, IL-15, and IL-18 Signaling to Induce Memory-Like NK Cells for Cancer Immunotherapy. Cancer Immunol. Res. 2021, 9, 1071–1087. [Google Scholar] [CrossRef]
- Cozar, B.; Greppi, M.; Carpentier, S.; Narni-Mancinelli, E.; Chiossone, L.; Vivier, E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021, 11, 34–44. [Google Scholar] [CrossRef]
- Bi, J.; Tian, Z. NK Cell Dysfunction and Checkpoint Immunotherapy. Front. Immunol. 2019, 10, 1999. [Google Scholar] [CrossRef]
- Boles, K.S.; Barten, R.; Kumaresan, P.R.; Trowsdale, J.; Mathew, P.A. Cloning of a new lectin-like receptor expressed on human NK cells. Immunogenetics 1999, 50, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Boles, K.S.; Mathew, P.A. Molecular cloning of CS1, a novel human natural killer cell receptor belonging to the CD2 subset of the immunoglobulin superfamily. Immunogenetics 2001, 52, 302–307. [Google Scholar] [CrossRef]
- Boles, K.S.; Nakajima, H.; Colonna, M.; Chuang, S.S.; Stepp, S.E.; Bennett, M.; Kumar, V.; Mathew, P.A. Molecular characterization of a novel human natural killer cell receptor homologous to mouse 2B4. Tissue Antigens 1999, 54, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Mathew, S.O.; Patel, R.K.; Pertusi, R.M.; Mathew, P.A. Altered expression of signalling lymphocyte activation molecule (SLAM) family receptors CS1 (CD319) and 2B4 (CD244) in patients with systemic lupus erythematosus. Clin. Exp. Immunol. 2010, 160, 348–358. [Google Scholar] [CrossRef]
- Mathew, S.O.; Chaudhary, P.; Powers, S.B.; Vishwanatha, J.K.; Mathew, P.A. Overexpression of LLT1 (OCIL, CLEC2D) on prostate cancer cells inhibits NK cell-mediated killing through LLT1-NKRP1A (CD161) interaction. Oncotarget 2016, 7, 68650–68661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrufo, A.M.; Mathew, S.O.; Chaudhary, P.; Malaer, J.D.; Vishwanatha, J.K.; Mathew, P.A. Blocking LLT1 (CLEC2D, OCIL)-NKRP1A (CD161) interaction enhances natural killer cell-mediated lysis of triple-negative breast cancer cells. Am. J. Cancer Res. 2018, 8, 1050–1063. [Google Scholar] [PubMed]
- Malaer, J.D.; Mathew, P.A. CS1 (SLAMF7, CD319) is an effective immunotherapeutic target for multiple myeloma. Am. J. Cancer Res. 2017, 7, 1637–1641. [Google Scholar] [PubMed]
- Buller, C.W.; Mathew, P.A.; Mathew, S.O. Roles of NK Cell Receptors 2B4 (CD244), CS1 (CD319), and LLT1 (CLEC2D) in Cancer. Cancers 2020, 12, 1755. [Google Scholar] [CrossRef] [PubMed]
- Malaer, J.D.; Marrufo, A.M.; Mathew, P.A. 2B4 (CD244, SLAMF4) and CS1 (CD319, SLAMF7) in systemic lupus erythematosus and cancer. Clin. Immunol. 2019, 204, 50–56. [Google Scholar] [CrossRef]
- Barrow, A.D.; Martin, C.J.; Colonna, M. The Natural Cytotoxicity Receptors in Health and Disease. Front. Immunol. 2019, 10, 909. [Google Scholar] [CrossRef] [Green Version]
- Hecht, M.L.; Rosental, B.; Horlacher, T.; Hershkovitz, O.; De Paz, J.L.; Noti, C.; Schauer, S.; Porgador, A.; Seeberger, P.H. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J. Proteome Res. 2009, 8, 712–720. [Google Scholar] [CrossRef]
- Campos, C.; Lopez, N.; Pera, A.; Gordillo, J.J.; Hassouneh, F.; Tarazona, R.; Solana, R. Expression of NKp30, NKp46 and DNAM-1 activating receptors on resting and IL-2 activated NK cells from healthy donors according to CMV-serostatus and age. Biogerontology 2015, 16, 671–683. [Google Scholar] [CrossRef]
- Nguyen, K.; Devidas, M.; Cheng, S.C.; La, M.; Raetz, E.A.; Carroll, W.L.; Winick, N.J.; Hunger, S.P.; Gaynon, P.S.; Loh, M.L.; et al. Factors influencing survival after relapse from acute lymphoblastic leukemia: A Children’s Oncology Group study. Leukemia 2008, 22, 2142–2150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallen, G.; Ratei, R.; Mann, G.; Kaspers, G.; Niggli, F.; Karachunsky, A.; Ebell, W.; Escherich, G.; Schrappe, M.; Klingebiel, T.; et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: Results of trial ALL-REZ BFM 90. J. Clin. Oncol. 2010, 28, 2339–2347. [Google Scholar] [CrossRef]
- Oskarsson, T.; Soderhall, S.; Arvidson, J.; Forestier, E.; Montgomery, S.; Bottai, M.; Lausen, B.; Carlsen, N.; Hellebostad, M.; Lahteenmaki, P.; et al. Relapsed childhood acute lymphoblastic leukemia in the Nordic countries: Prognostic factors, treatment and outcome. Haematologica 2016, 101, 68–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceppi, F.; Cazzaniga, G.; Colombini, A.; Biondi, A.; Conter, V. Risk factors for relapse in childhood acute lymphoblastic leukemia: Prediction and prevention. Expert Rev. Hematol. 2015, 8, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H. Precision medicine in acute lymphoblastic leukemia. Front. Med. 2020, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Maciocia, P.M.; Wawrzyniecka, P.A.; Maciocia, N.C.; Burley, A.; Karpanasamy, T.; Devereaux, S.; Hoekx, M.; O’Connor, D.; Leon, T.; Rapoz-D’Silva, T.; et al. Anti-CCR9 chimeric antigen receptor T cells for T-cell acute lymphoblastic leukemia. Blood 2022, 140, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Kadia, T.M.; Gandhi, V. Nelarabine in the treatment of pediatric and adult patients with T-cell acute lymphoblastic leukemia and lymphoma. Expert Rev. Hematol. 2017, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Chester, C.; Fritsch, K.; Kohrt, H.E. Natural Killer Cell Immunomodulation: Targeting Activating, Inhibitory, and Co-stimulatory Receptor Signaling for Cancer Immunotherapy. Front. Immunol. 2015, 6, 601. [Google Scholar] [CrossRef] [Green Version]
- Rosen, D.B.; Bettadapura, J.; Alsharifi, M.; Mathew, P.A.; Warren, H.S.; Lanier, L.L. Cutting edge: Lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J. Immunol. 2005, 175, 7796–7799. [Google Scholar] [CrossRef] [Green Version]
- Roth, P.; Mittelbronn, M.; Wick, W.; Meyermann, R.; Tatagiba, M.; Weller, M. Malignant glioma cells counteract antitumor immune responses through expression of lectin-like transcript-1. Cancer Res. 2007, 67, 3540–3544. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Canteli, M.; Hermida-Prado, F.; Sordo-Bahamonde, C.; Montoro-Jimenez, I.; Pozo-Agundo, E.; Allonca, E.; Vallina-Alvarez, A.; Alvarez-Marcos, C.; Gonzalez, S.; Garcia-Pedrero, J.M.; et al. Lectin-Like Transcript 1 (LLT1) Checkpoint: A Novel Independent Prognostic Factor in HPV-Negative Oropharyngeal Squamous Cell Carcinoma. Biomedicines 2020, 8, 535. [Google Scholar] [CrossRef]
- Malaer, J.D.; Mathew, P.A. Role of LLT1 and PCNA as Natural Killer Cell Immune Evasion Strategies of HCT 116 Cells. Anticancer Res. 2020, 40, 6613–6621. [Google Scholar] [CrossRef]
- Braud, V.M.; Biton, J.; Becht, E.; Knockaert, S.; Mansuet-Lupo, A.; Cosson, E.; Damotte, D.; Alifano, M.; Validire, P.; Anjuere, F.; et al. Expression of LLT1 and its receptor CD161 in lung cancer is associated with better clinical outcome. Oncoimmunology 2018, 7, e1423184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Germain, C.; Guillaudeux, T.; Galsgaard, E.D.; Hervouet, C.; Tekaya, N.; Gallouet, A.S.; Fassy, J.; Bihl, F.; Poupon, G.; Lazzari, A.; et al. Lectin-like transcript 1 is a marker of germinal center-derived B-cell non-Hodgkin’s lymphomas dampening natural killer cell functions. Oncoimmunology 2015, 4, e1026503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braud, V.M.; Meghraoui-Kheddar, A.; Elaldi, R.; Petti, L.; Germain, C.; Anjuere, F. LLT1-CD161 Interaction in Cancer: Promises and Challenges. Front. Immunol. 2022, 13, 847576. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Tian, Z. NK Cell Exhaustion. Front. Immunol. 2017, 8, 760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Judge, S.J.; Murphy, W.J.; Canter, R.J. Characterizing the Dysfunctional NK Cell: Assessing the Clinical Relevance of Exhaustion, Anergy, and Senescence. Front. Cell. Infect. Microbiol. 2020, 10, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsi, E.D.; Steinle, R.; Balasa, B.; Szmania, S.; Draksharapu, A.; Shum, B.P.; Huseni, M.; Powers, D.; Nanisetti, A.; Zhang, Y.; et al. CS1, a potential new therapeutic antibody target for the treatment of multiple myeloma. Clin. Cancer Res. 2008, 14, 2775–2784. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.R.; Horton, N.C.; Mathew, S.O.; Mathew, P.A. CS1 (SLAMF7) inhibits production of proinflammatory cytokines by activated monocytes. Inflamm. Res. 2013, 62, 765–772. [Google Scholar] [CrossRef]
- O’Connell, P.; Pepelyayeva, Y.; Blake, M.K.; Hyslop, S.; Crawford, R.B.; Rizzo, M.D.; Pereira-Hicks, C.; Godbehere, S.; Dale, L.; Gulick, P.; et al. SLAMF7 Is a Critical Negative Regulator of IFN-alpha-Mediated CXCL10 Production in Chronic HIV Infection. J. Immunol. 2019, 202, 228–238. [Google Scholar] [CrossRef] [Green Version]
- Maekawa, T.; Kato, S.; Kawamura, T.; Takada, K.; Sone, T.; Ogata, H.; Saito, K.; Izumi, T.; Nagao, S.; Takano, K.; et al. Increased SLAMF7(high) monocytes in myelofibrosis patients harboring JAK2V617F provide a therapeutic target of elotuzumab. Blood 2019, 134, 814–825. [Google Scholar] [CrossRef]
- Chalan, P.; Bijzet, J.; Huitema, M.G.; Kroesen, B.J.; Brouwer, E.; Boots, A.M. Expression of Lectin-Like Transcript 1, the Ligand for CD161, in Rheumatoid Arthritis. PLoS ONE 2015, 10, e0132436. [Google Scholar] [CrossRef]
- Schlaphoff, V.; Lunemann, S.; Suneetha, P.V.; Jaroszewicz, J.; Grabowski, J.; Dietz, J.; Helfritz, F.; Bektas, H.; Sarrazin, C.; Manns, M.P.; et al. Dual function of the NK cell receptor 2B4 (CD244) in the regulation of HCV-specific CD8+ T cells. PLoS Pathog. 2011, 7, e1002045. [Google Scholar] [CrossRef]
- Boieri, M.; Ulvmoen, A.; Sudworth, A.; Lendrem, C.; Collin, M.; Dickinson, A.M.; Kveberg, L.; Inngjerdingen, M. IL-12, IL-15, and IL-18 pre-activated NK cells target resistant T cell acute lymphoblastic leukemia and delay leukemia development in vivo. Oncoimmunology 2017, 6, e1274478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Iglesias, T.; Del Toro-Arreola, A.; Albarran-Somoza, B.; Del Toro-Arreola, S.; Sanchez-Hernandez, P.E.; Ramirez-Duenas, M.G.; Balderas-Pena, L.M.; Bravo-Cuellar, A.; Ortiz-Lazareno, P.C.; Daneri-Navarro, A. Low NKp30, NKp46 and NKG2D expression and reduced cytotoxic activity on NK cells in cervical cancer and precursor lesions. BMC Cancer 2009, 9, 186. [Google Scholar] [CrossRef]


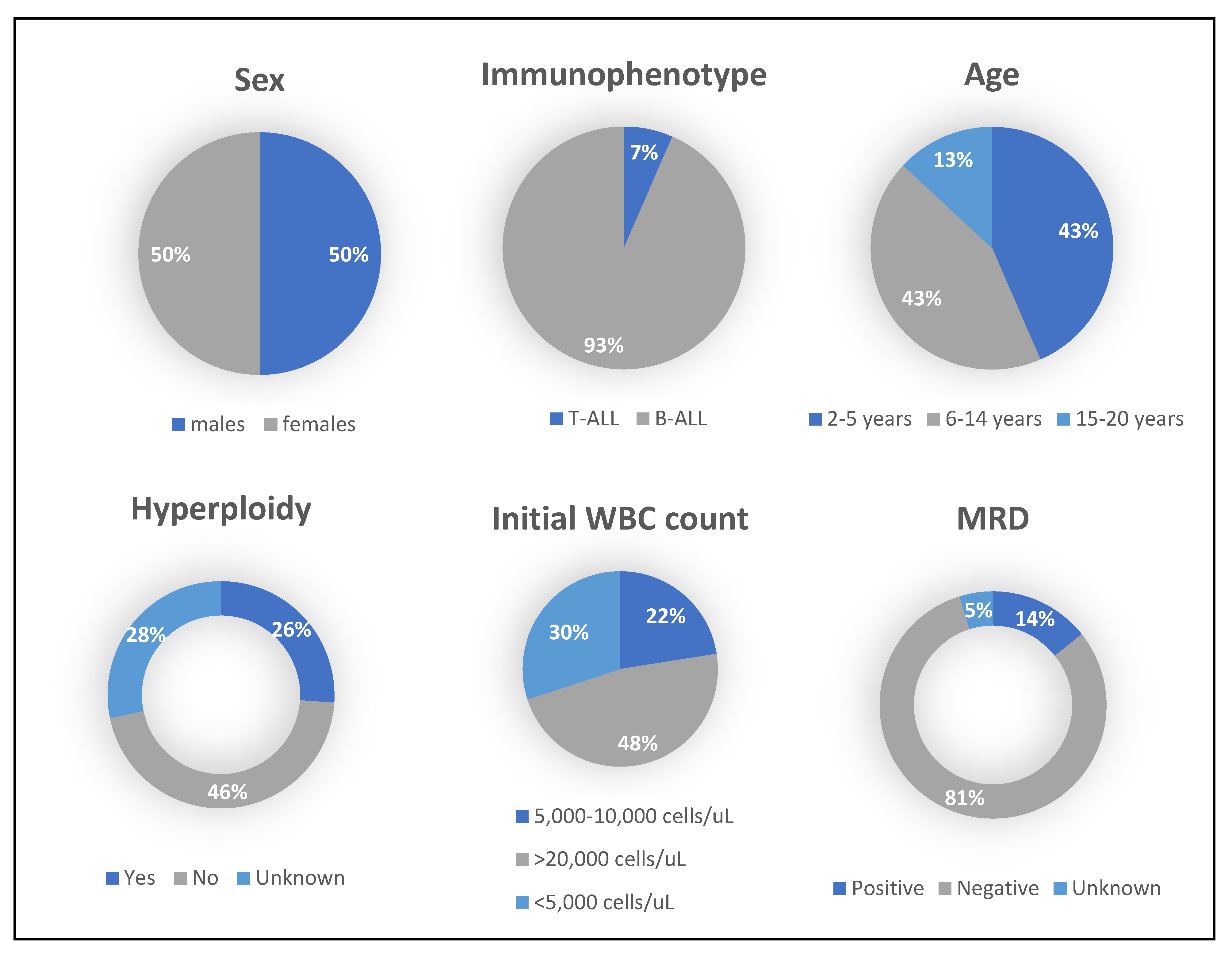
| Clinical Features | No. of Patients |
|---|---|
| Sex | |
| Male | 21 |
| Female | 21 |
| ALL Subtype | |
| B-ALL | 39 |
| T-ALL | 3 |
| Age | |
| 2–5 | 18 |
| 6–14 | 18 |
| 15–20 | 6 |
| MRD | |
| Positive | 6 |
| Negative | 34 |
| Unknown | 2 |
| Initial WBC count (Cells/ul) | |
| <5000 | 12 |
| 5000–10,000 | 9 |
| >20,000 | 19 |
| Hyperdiploidy | |
| Yes | 12 |
| No | 19 |
| Unknown | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Powers, S.B.; Ahmed, N.G.; Jose, R.; Brezgiel, M.; Aryal, S.; Bowman, W.P.; Mathew, P.A.; Mathew, S.O. Differential Expression of LLT1, SLAM Receptors CS1 and 2B4 and NCR Receptors NKp46 and NKp30 in Pediatric Acute Lymphoblastic Leukemia (ALL). Int. J. Mol. Sci. 2023, 24, 3860. https://doi.org/10.3390/ijms24043860
Powers SB, Ahmed NG, Jose R, Brezgiel M, Aryal S, Bowman WP, Mathew PA, Mathew SO. Differential Expression of LLT1, SLAM Receptors CS1 and 2B4 and NCR Receptors NKp46 and NKp30 in Pediatric Acute Lymphoblastic Leukemia (ALL). International Journal of Molecular Sciences. 2023; 24(4):3860. https://doi.org/10.3390/ijms24043860
Chicago/Turabian StylePowers, Sheila B., Nourhan G. Ahmed, Roslin Jose, Marissa Brezgiel, Subhash Aryal, W. Paul Bowman, Porunelloor A. Mathew, and Stephen O. Mathew. 2023. "Differential Expression of LLT1, SLAM Receptors CS1 and 2B4 and NCR Receptors NKp46 and NKp30 in Pediatric Acute Lymphoblastic Leukemia (ALL)" International Journal of Molecular Sciences 24, no. 4: 3860. https://doi.org/10.3390/ijms24043860





