Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions
Abstract
:1. Introduction
2. Results
2.1. Obesity-Related CircRNAs Were Specifically Expressed in Brain Cells
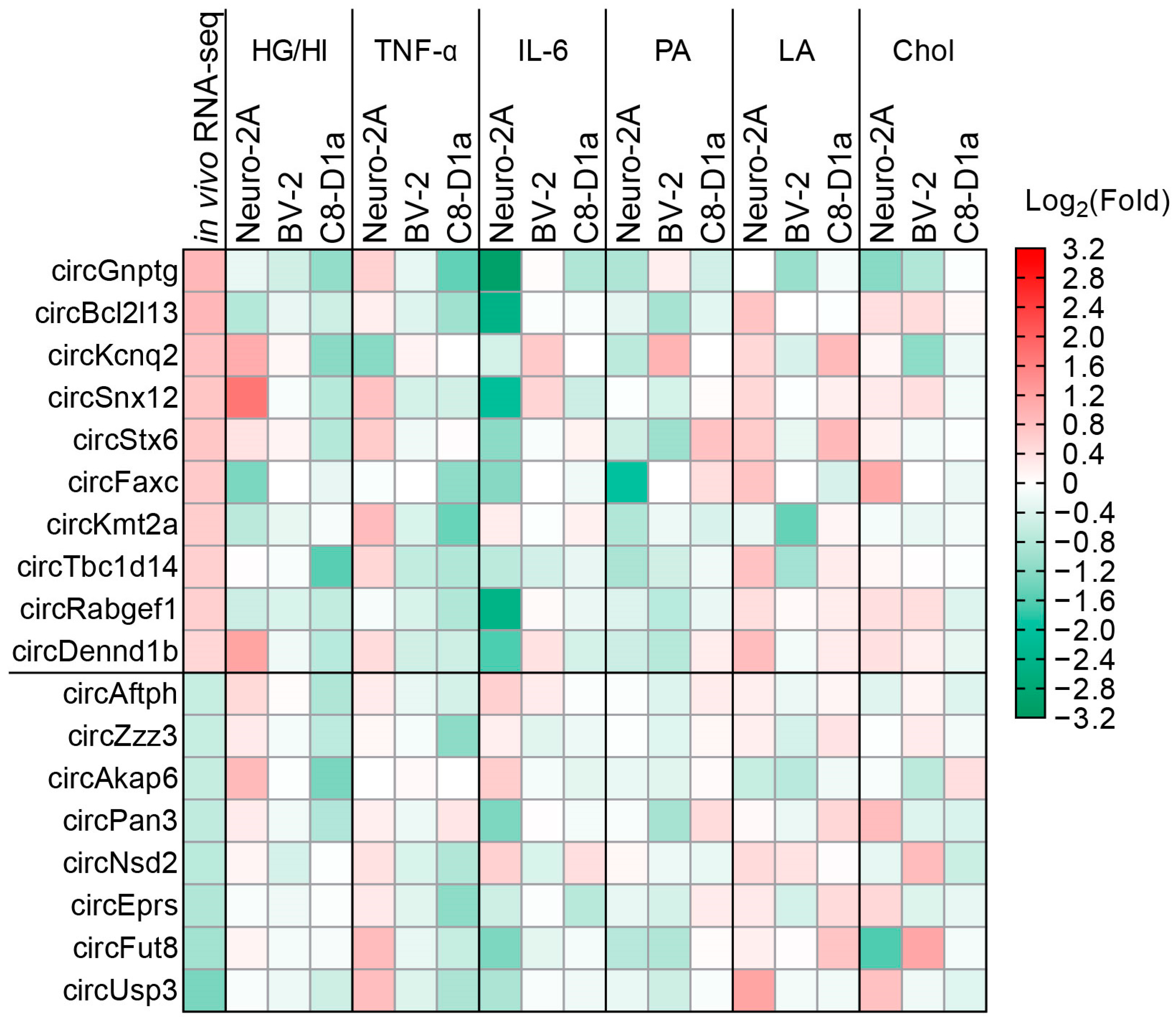
2.2. Obesity-Related In Vitro Conditions Regulated the Expression of CircRNAs in Brain Cells
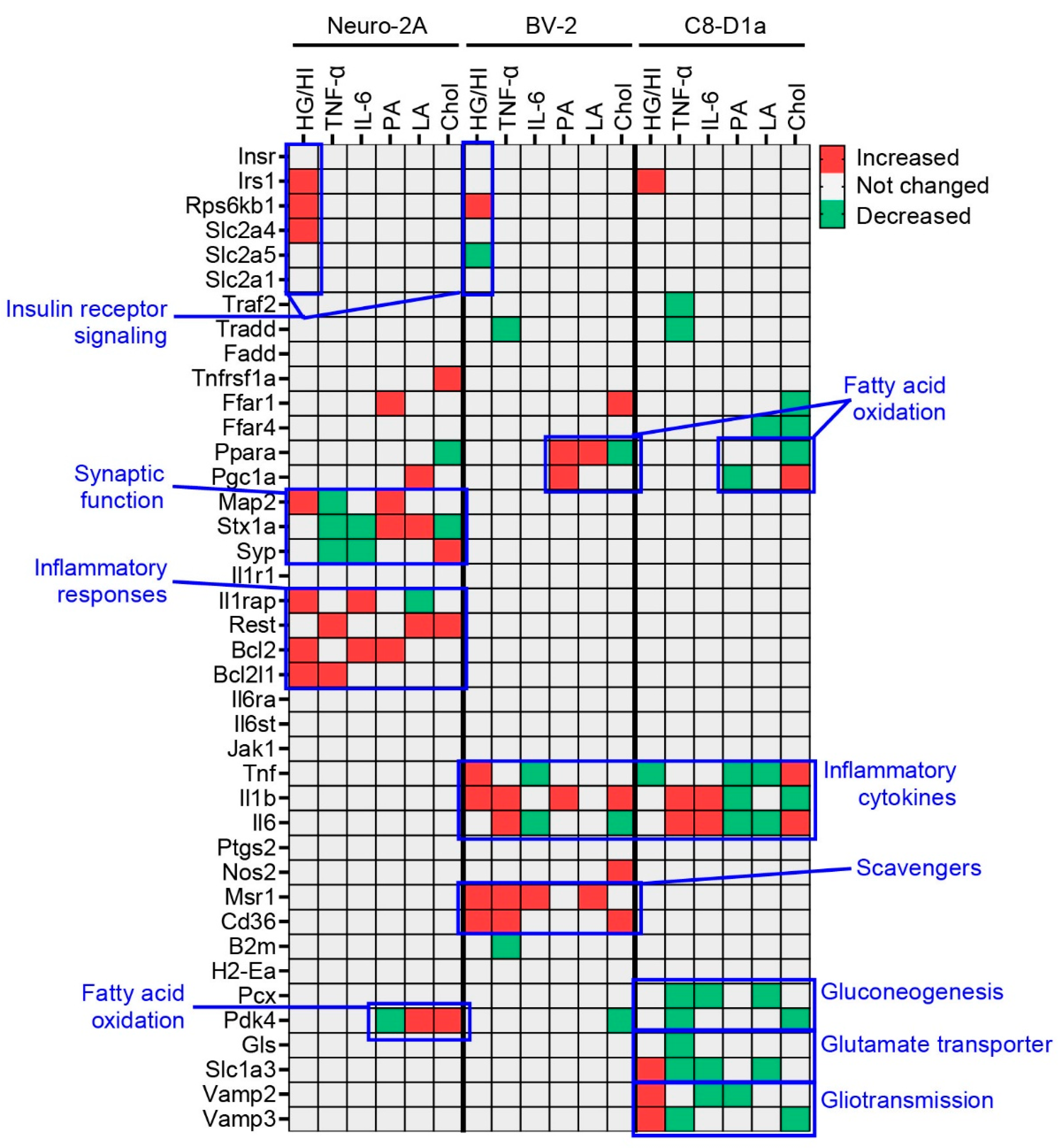
2.3. Obesity-Related In Vitro Conditions Affected the Cell Type-Specific Functions of Brain Cells
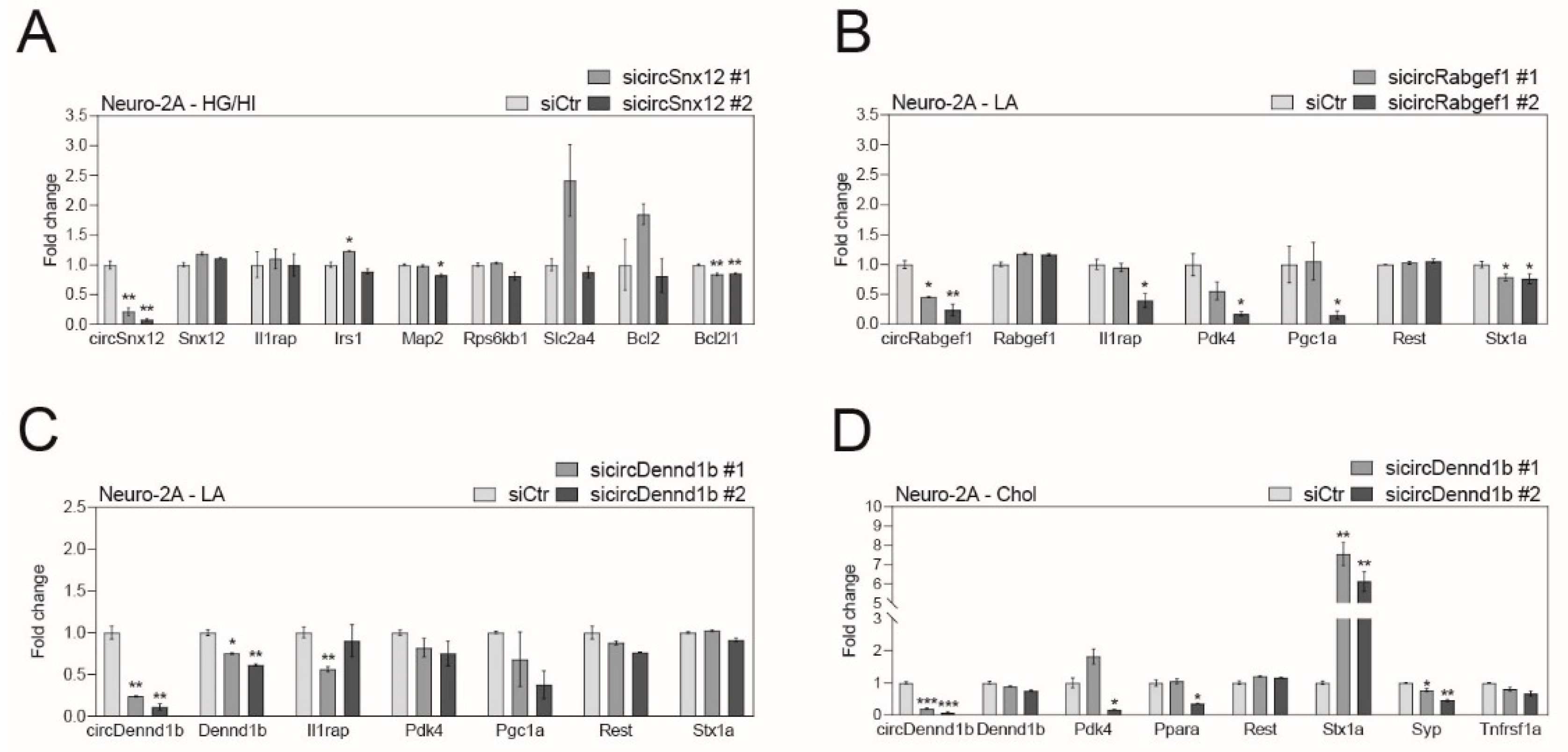
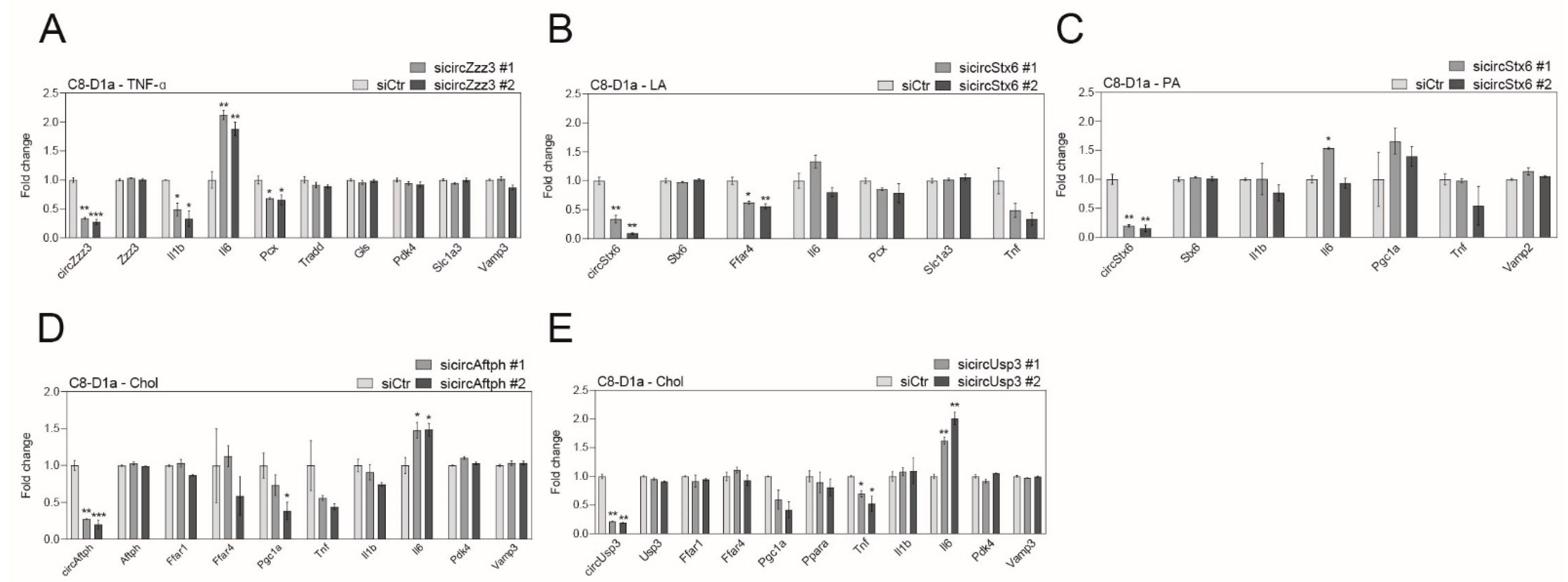
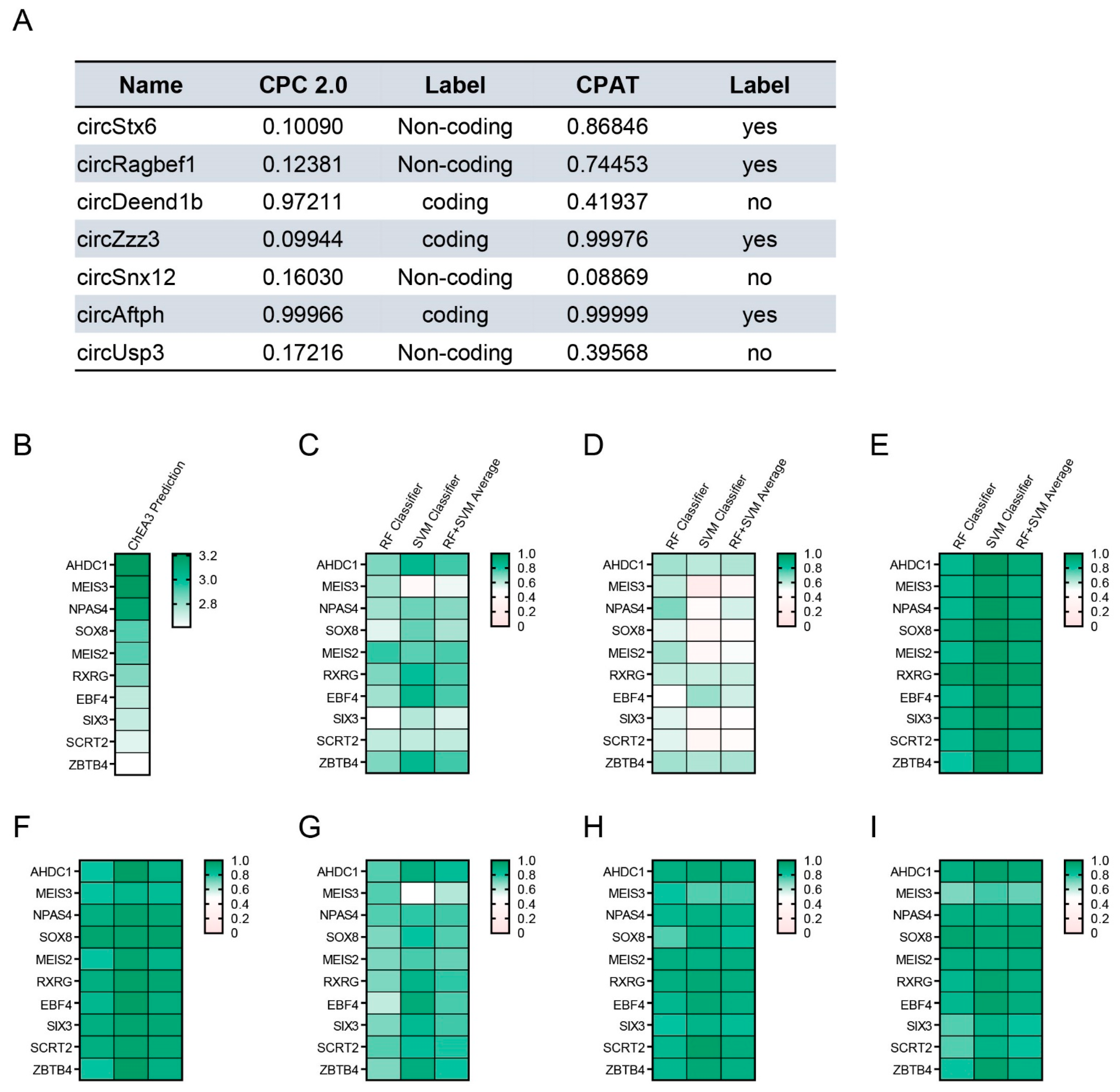
2.4. Obesity-Related CircRNAs Work as Important Factors for Brain Cell Function
2.5. Analysis of the Protein Interaction for Obesity-Related CircRNAs
3. Discussion
4. Materials and Methods
4.1. Drug Treatment
4.2. Cell line and Culture Conditions
4.3. siRNA Design and Transfection
4.4. RNA Isolation and Semi-Quantitative Polymerase Chain Reaction
4.5. Confirmation of the Circular Structure of the CircRNAs
4.6. Analysis of CircRNA Function
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knopman, D.S.; Amieva, H.; Petersen, R.C.; Chetelat, G.; Holtzman, D.M.; Hyman, B.T.; Nixon, R.A.; Jones, D.T. Alzheimer disease. Nat. Rev. Dis. Prim. 2021, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Selkoe, D.J. The amyloid hypothesis of Alzheimer’s disease: Progress and problems on the road to therapeutics. Science 2002, 297, 353–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hardy, J.A.; Higgins, G.A. Alzheimer’s disease: The amyloid cascade hypothesis. Science 1992, 256, 184–185. [Google Scholar] [CrossRef] [PubMed]
- Arnsten, A.F.T.; Datta, D.; Del Tredici, K.; Braak, H. Hypothesis: Tau pathology is an initiating factor in sporadic Alzheimer’s disease. Alzheimers Dement. 2021, 17, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; De Strooper, B.; Kivipelto, M.; Holstege, H.; Chetelat, G.; Teunissen, C.E.; Cummings, J.; van der Flier, W.M. Alzheimer’s disease. Lancet 2021, 397, 1577–1590. [Google Scholar] [CrossRef]
- Rios, J.A.; Cisternas, P.; Arrese, M.; Barja, S.; Inestrosa, N.C. Is Alzheimer’s disease related to metabolic syndrome? A Wnt signaling conundrum. Prog. Neurobiol. 2014, 121, 125–146. [Google Scholar] [CrossRef] [PubMed]
- Biessels, G.J.; Despa, F. Cognitive decline and dementia in diabetes mellitus: Mechanisms and clinical implications. Nat. Rev. Endocrinol. 2018, 14, 591–604. [Google Scholar] [CrossRef]
- Li, T.; Qu, J.; Xu, C.; Fang, T.; Sun, B.; Chen, L. Exploring the common gene signatures and pathogeneses of obesity with Alzheimer’s disease via transcriptome data. Front. Endocrinol. 2022, 13, 1072955. [Google Scholar] [CrossRef]
- Matsuzaki, T.; Sasaki, K.; Tanizaki, Y.; Hata, J.; Fujimi, K.; Matsui, Y.; Sekita, A.; Suzuki, S.O.; Kanba, S.; Kiyohara, Y.; et al. Insulin resistance is associated with the pathology of Alzheimer disease: The Hisayama study. Neurology 2010, 75, 764–770. [Google Scholar] [CrossRef]
- Chen, H.; Du, Y.; Liu, S.; Ge, B.; Ji, Y.; Huang, G. Association between serum cholesterol levels and Alzheimer’s disease in China: A case-control study. Int. J. Food Sci. Nutr. 2019, 70, 405–411. [Google Scholar] [CrossRef]
- Wang, D.C.; Sun, C.H.; Liu, L.Y.; Sun, X.H.; Jin, X.W.; Song, W.L.; Liu, X.Q.; Wan, X.L. Serum fatty acid profiles using GC-MS and multivariate statistical analysis: Potential biomarkers of Alzheimer’s disease. Neurobiol. Aging 2012, 33, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Long, C.; Hu, X.; Chai, X. Obesity is associated with greater cognitive function in patients with type 2 diabetes mellitus. Front. Endocrinol. 2022, 13, 953826. [Google Scholar] [CrossRef] [PubMed]
- Zanini, P.; Arbo, B.D.; Niches, G.; Czarnabay, D.; Benetti, F.; Ribeiro, M.F.; Cecconello, A.L. Diet-induced obesity alters memory consolidation in female rats. Physiol. Behav. 2017, 180, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Sedzikowska, A.; Szablewski, L. Insulin and Insulin Resistance in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 9987. [Google Scholar] [CrossRef]
- Haas, C.B.; de Carvalho, A.K.; Muller, A.P.; Eggen, B.J.L.; Portela, L.V. Insulin activates microglia and increases COX-2/IL-1beta expression in young but not in aged hippocampus. Brain Res. 2020, 1741, 146884. [Google Scholar] [CrossRef]
- Heni, M.; Eckstein, S.S.; Schittenhelm, J.; Bohm, A.; Hogrefe, N.; Irmler, M.; Beckers, J.; Hrabe de Angelis, M.; Haring, H.U.; Fritsche, A.; et al. Ectopic fat accumulation in human astrocytes impairs insulin action. R. Soc. Open Sci. 2020, 7, 200701. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, J.; Jiang, W.; Cao, Z.; Zhao, F.; Cai, T.; Aschner, M.; Luo, W. The role of NLRP3-CASP1 in inflammasome-mediated neuroinflammation and autophagy dysfunction in manganese-induced, hippocampal-dependent impairment of learning and memory ability. Autophagy 2017, 13, 914–927. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Yang, S.; Luo, C. TNF-alpha and TNF-R1 regulate bupivacaine-induced apoptosis in spinal cord dorsal root ganglion neuron. Eur. J. Pharmacol. 2018, 833, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Russ, K.; Teku, G.; Bousset, L.; Redeker, V.; Piel, S.; Savchenko, E.; Pomeshchik, Y.; Savistchenko, J.; Stummann, T.C.; Azevedo, C.; et al. TNF-alpha and alpha-synuclein fibrils differently regulate human astrocyte immune reactivity and impair mitochondrial respiration. Cell Rep. 2021, 34, 108895. [Google Scholar] [CrossRef]
- Spinelli, M.; Fusco, S.; Mainardi, M.; Scala, F.; Natale, F.; Lapenta, R.; Mattera, A.; Rinaudo, M.; Li Puma, D.D.; Ripoli, C.; et al. Brain insulin resistance impairs hippocampal synaptic plasticity and memory by increasing GluA1 palmitoylation through FoxO3a. Nat. Commun. 2017, 8, 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Alegria, K.; Bastian-Eugenio, C.E.; Vaca, L.; Arias, C. Palmitic acid induces insulin resistance by a mechanism associated with energy metabolism and calcium entry in neuronal cells. FASEB J. 2021, 35, e21712. [Google Scholar] [CrossRef]
- Kulas, J.A.; Weigel, T.K.; Ferris, H.A. Insulin resistance and impaired lipid metabolism as a potential link between diabetes and Alzheimer’s disease. Drug Dev. Res. 2020, 81, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Liu, S.; Lopes-Virella, M.F.; Wang, Z. LPS and palmitic acid Co-upregulate microglia activation and neuroinflammatory response. Compr. Psychoneuroendocrinol. 2021, 6, 100048. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Rodriguez, A.; Acaz-Fonseca, E.; Boya, P.; Arevalo, M.A.; Garcia-Segura, L.M. Lipotoxic Effects of Palmitic Acid on Astrocytes Are Associated with Autophagy Impairment. Mol. Neurobiol. 2019, 56, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Takousis, P.; Sadlon, A.; Schulz, J.; Wohlers, I.; Dobricic, V.; Middleton, L.; Lill, C.M.; Perneczky, R.; Bertram, L. Differential expression of microRNAs in Alzheimer’s disease brain, blood, and cerebrospinal fluid. Alzheimers Dement. 2019, 15, 1468–1477. [Google Scholar] [CrossRef]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- You, X.; Vlatkovic, I.; Babic, A.; Will, T.; Epstein, I.; Tushev, G.; Akbalik, G.; Wang, M.; Glock, C.; Quedenau, C.; et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015, 18, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Yoon, G.; Cho, K.A.; Song, J.; Kim, Y.K. Transcriptomic Analysis of High Fat Diet Fed Mouse Brain Cortex. Front. Genet. 2019, 10, 83. [Google Scholar] [CrossRef] [Green Version]
- Yoon, G.; Lim, Y.H.; Jo, D.; Ryu, J.; Song, J.; Kim, Y.K. Obesity-linked circular RNA circTshz2-2 regulates the neuronal cell cycle and spatial memory in the brain. Mol. Psychiatry 2021, 26, 6350–6364. [Google Scholar] [CrossRef] [PubMed]
- Patil, N.S.; Feng, B.; Su, Z.; Castellani, C.A.; Chakrabarti, S. Circular RNA mediated gene regulation in chronic diabetic complications. Sci. Rep. 2021, 11, 23766. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zheng, G.; Ning, Q.; Zheng, J.; Dong, D. Translation and functional roles of circular RNAs in human cancer. Mol. Cancer 2020, 19, 30. [Google Scholar] [CrossRef] [Green Version]
- Shan, K.; Liu, C.; Liu, B.H.; Chen, X.; Dong, R.; Liu, X.; Zhang, Y.Y.; Liu, B.; Zhang, S.J.; Wang, J.J.; et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017, 136, 1629–1642. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yoon, G.; Song, J.; Jo, J. Exendin-4 improves long-term potentiation and neuronal dendritic growth in vivo and in vitro obesity condition. Sci. Rep. 2021, 11, 8326. [Google Scholar] [CrossRef] [PubMed]
- Yoon, G.; Kim, Y.K.; Song, J. Glucagon-like peptide-1 suppresses neuroinflammation and improves neural structure. Pharmacol. Res. 2020, 152, 104615. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Li, X.; Cao, P.; Fei, W.; Zhou, H.; Tang, N.; Liu, Y. Interleukin-6 mediated inflammasome activation promotes oral squamous cell carcinoma progression via JAK2/STAT3/Sox4/NLRP3 signaling pathway. J. Exp. Clin. Cancer Res. 2022, 41, 166. [Google Scholar] [CrossRef]
- Jo, D.; Yoon, G.; Song, J. Role of Exendin-4 in Brain Insulin Resistance, Mitochondrial Function, and Neurite Outgrowth in Neurons under Palmitic Acid-Induced Oxidative Stress. Antioxidants 2021, 10, 78. [Google Scholar] [CrossRef]
- Reardon, M.; Gobern, S.; Martinez, K.; Shen, W.; Reid, T.; McIntosh, M. Oleic acid attenuates trans-10,cis-12 conjugated linoleic acid-mediated inflammatory gene expression in human adipocytes. Lipids 2012, 47, 1043–1051. [Google Scholar] [CrossRef] [Green Version]
- Avila-Munoz, E.; Arias, C. Cholesterol-induced astrocyte activation is associated with increased amyloid precursor protein expression and processing. Glia 2015, 63, 2010–2022. [Google Scholar] [CrossRef]
- Ryu, J.; Choe, N.; Kwon, D.H.; Shin, S.; Lim, Y.H.; Yoon, G.; Kim, J.H.; Kim, H.S.; Lee, I.K.; Ahn, Y.; et al. Circular RNA circSmoc1-2 regulates vascular calcification by acting as a miR-874-3p sponge in vascular smooth muscle cells. Mol. Ther. Nucleic Acids 2022, 27, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Keenan, A.B.; Torre, D.; Lachmann, A.; Leong, A.K.; Wojciechowicz, M.L.; Utti, V.; Jagodnik, K.M.; Kropiwnicki, E.; Wang, Z.; Ma’ayan, A. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019, 47, W212–W224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muppirala, U.K.; Honavar, V.G.; Dobbs, D. Predicting RNA-protein interactions using only sequence information. BMC Bioinform. 2011, 12, 489. [Google Scholar] [CrossRef] [Green Version]
- van Heesch, S.; Witte, F.; Schneider-Lunitz, V.; Schulz, J.F.; Adami, E.; Faber, A.B.; Kirchner, M.; Maatz, H.; Blachut, S.; Sandmann, C.L.; et al. The Translational Landscape of the Human Heart. Cell 2019, 178, 242–260.e229. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.; Wang, S.K.; Belk, J.A.; Amaya, L.; Li, Z.; Cardenas, A.; Abe, B.T.; Chen, C.K.; Wender, P.A.; Chang, H.Y. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 2022, 41, 262–272. [Google Scholar] [CrossRef]
- Kim, S.; Park, D.; Kim, J.; Kim, D.; Kim, H.; Mori, T.; Jung, H.; Lee, D.; Hong, S.; Jeon, J.; et al. Npas4 regulates IQSEC3 expression in hippocampal somatostatin interneurons to mediate anxiety-like behavior. Cell. Rep. 2021, 36, 109417. [Google Scholar] [CrossRef]
- Takouda, J.; Katada, S.; Imamura, T.; Sanosaka, T.; Nakashima, K. SoxE group transcription factor Sox8 promotes astrocytic differentiation of neural stem/precursor cells downstream of Nfia. Pharmacol. Res. Perspect. 2021, 9, e00749. [Google Scholar] [CrossRef] [PubMed]
- Wietrzych, M.; Meziane, H.; Sutter, A.; Ghyselinck, N.; Chapman, P.F.; Chambon, P.; Krezel, W. Working memory deficits in retinoid X receptor gamma-deficient mice. Learn. Mem. 2005, 12, 318–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upreti, C.; Woodruff, C.M.; Zhang, X.L.; Yim, M.J.; Zhou, Z.Y.; Pagano, A.M.; Rehanian, D.S.; Yin, D.; Kandel, E.R.; Stanton, P.K.; et al. Loss of retinoid X receptor gamma subunit impairs group 1 mGluR mediated electrophysiological responses and group 1 mGluR dependent behaviors. Sci. Rep. 2021, 11, 5552. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, J.; Cao, X.; Cai, Z.; Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 2022, 13, 3242. [Google Scholar] [CrossRef]
- Xu, F.; Shen, L.; Chen, H.; Wang, R.; Zang, T.; Qian, J.; Ge, J. circDENND1B Participates in the Antiatherosclerotic Effect of IL-1beta Monoclonal Antibody in Mouse by Promoting Cholesterol Efflux via miR-17-5p/Abca1 Axis. Front. Cell. Dev Biol. 2021, 9, 652032. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, A.; Makino, N.; Tozawa, T.; Shirahata, N.; Honda, T.; Ikeda, Y.; Sato, H.; Ito, M.; Kakizaki, Y.; Akamatsu, M.; et al. Pancreatic fat accumulation, fibrosis, and acinar cell injury in the Zucker diabetic fatty rat fed a chronic high-fat diet. Pancreas 2014, 43, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Ludgero-Correia, A., Jr.; Aguila, M.B.; Mandarim-de-Lacerda, C.A.; Faria, T.S. Effects of high-fat diet on plasma lipids, adiposity, and inflammatory markers in ovariectomized C57BL/6 mice. Nutrition 2012, 28, 316–323. [Google Scholar] [CrossRef] [Green Version]
- Zhukova, N.V.; Novgorodtseva, T.P.; Denisenko, Y.K. Effect of the prolonged high-fat diet on the fatty acid metabolism in rat blood and liver. Lipids Health Dis. 2014, 13, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberhauser, F.; Schulte, D.M.; Faust, M.; Gudelhofer, H.; Hahn, M.; Muller, N.; Neumann, K.; Krone, W.; Laudes, M. Weight loss due to a very low calorie diet differentially affects insulin sensitivity and interleukin-6 serum levels in nondiabetic obese human subjects. Horm. Metab. Res. 2012, 44, 465–470. [Google Scholar] [CrossRef]
- Miszta, A.; Kopec, A.K.; Pant, A.; Holle, L.A.; Byrnes, J.R.; Lawrence, D.A.; Hansen, K.C.; Flick, M.J.; Luyendyk, J.P.; de Laat, B.; et al. A high-fat diet delays plasmin generation in a thrombomodulin-dependent manner in mice. Blood 2020, 135, 1704–1717. [Google Scholar] [CrossRef]
- Benson, T.W.; Weintraub, N.L.; Kim, H.W.; Seigler, N.; Kumar, S.; Pye, J.; Horimatsu, T.; Pellenberg, R.; Stepp, D.W.; Lucas, R.; et al. A single high-fat meal provokes pathological erythrocyte remodeling and increases myeloperoxidase levels: Implications for acute coronary syndrome. Lab. Invest. 2018, 98, 1300–1310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Jia, Y.; Guo, Y.; Zhao, N. Ion Migration in Perovskite Light-Emitting Diodes: Mechanism, Characterizations, and Material and Device Engineering. Adv. Mater. 2022, 34, e2108102. [Google Scholar] [CrossRef]
- Sheikh, M.H.; Errede, M.; d’Amati, A.; Khan, N.Q.; Fanti, S.; Loiola, R.A.; McArthur, S.; Purvis, G.S.D.; O’Riordan, C.E.; Ferorelli, D.; et al. Impact of metabolic disorders on the structural, functional, and immunological integrity of the blood-brain barrier: Therapeutic avenues. FASEB J. 2022, 36, e22107. [Google Scholar] [CrossRef] [PubMed]
- de Paula, G.C.; Brunetta, H.S.; Engel, D.F.; Gaspar, J.M.; Velloso, L.A.; Engblom, D.; de Oliveira, J.; de Bem, A.F. Hippocampal Function Is Impaired by a Short-Term High-Fat Diet in Mice: Increased Blood-Brain Barrier Permeability and Neuroinflammation as Triggering Events. Front. Neurosci. 2021, 15, 734158. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, F.; Mirzaie Bavil, F.; Keyhanmanesh, R.; Lotfi, H.; Ghiasi, F. Association of pro-inflammatory cytokines, inflammatory proteins with atherosclerosis index in obese male subjects. Horm. Mol. Biol. Clin. Investig. 2023. [Google Scholar] [CrossRef]
- Grao-Cruces, E.; Millan-Linares, M.C.; Martin-Rubio, M.E.; Toscano, R.; Barrientos-Trigo, S.; Bermudez, B.; Montserrat-de la Paz, S. Obesity-Associated Metabolic Disturbances Reverse the Antioxidant and Anti-Inflammatory Properties of High-Density Lipoproteins in Microglial Cells. Biomedicines 2021, 9, 1722. [Google Scholar] [CrossRef]
- Cleland, N.R.W.; Bruce, K.D. Fatty acid sensing in the brain: The role of glial-neuronal metabolic crosstalk and horizontal lipid flux. Biochimie 2022. [Google Scholar] [CrossRef]
- Chen, C.; Wen, M.; Wang, C.; Yuan, Z.; Jin, Y. Differential proteomic analysis of mouse cerebrums with high-fat diet (HFD)-induced hyperlipidemia. PeerJ 2022, 10, e13806. [Google Scholar] [CrossRef]
- Abd-Rabo, M.M.; Wahman, L.F.; El Hosary, R.; Ahmed, I.S. High-fat diet induced alteration in lipid enzymes and inflammation in cardiac and brain tissues: Assessment of the effects of Atorvastatin-loaded nanoparticles. J. Biochem. Mol. Toxicol. 2020, 34, e22465. [Google Scholar] [CrossRef]
- Howe, A.M.; Burke, S.; O’Reilly, M.E.; McGillicuddy, F.C.; Costello, D.A. Palmitic Acid and Oleic Acid Differently Modulate TLR2-Mediated Inflammatory Responses in Microglia and Macrophages. Mol. Neurobiol. 2022, 59, 2348–2362. [Google Scholar] [CrossRef]
- Pekkarinen, L.; Kantonen, T.; Rebelos, E.; Latva-Rasku, A.; Dadson, P.; Karjalainen, T.; Bucci, M.; Kalliokoski, K.; Laitinen, K.; Houttu, N.; et al. Obesity risk is associated with brain glucose uptake and insulin resistance. Eur. J. Endocrinol. 2022, 187, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, E.; Cetin-Tas, Y.; Sekerdag, E.; Yigit, B.; Shomalizadeh, N.; Sapanci, S.; Ozler, C.; Kesibi, J.; Kizilirmak, A.B.; Pekmez, M.; et al. Hyperglycemia with or without insulin resistance triggers different structural changes in brain microcirculation and perivascular matrix. Metab. Brain Dis. 2022, 38, 307–321. [Google Scholar] [CrossRef]
- Mamounis, K.J.; Yasrebi, A.; Roepke, T.A. Linoleic acid causes greater weight gain than saturated fat without hypothalamic inflammation in the male mouse. J. Nutr. Biochem. 2017, 40, 122–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Matute, P.; Martinez, J.A.; Marti, A.; Moreno-Aliaga, M.J. Linoleic acid decreases leptin and adiponectin secretion from primary rat adipocytes in the presence of insulin. Lipids 2007, 42, 913–920. [Google Scholar] [CrossRef]
- Pugazhenthi, S.; Qin, L.; Reddy, P.H. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 1037–1045. [Google Scholar] [CrossRef]
- Garcia, S.M.; Hirschberg, P.R.; Sarkar, P.; Siegel, D.M.; Teegala, S.B.; Vail, G.M.; Routh, V.H. Insulin actions on hypothalamic glucose-sensing neurones. J. Neuroendocrinol. 2021, 33, e12937. [Google Scholar] [CrossRef]
- Hunt, W.T.; Kamboj, A.; Anderson, H.D.; Anderson, C.M. Protection of cortical neurons from excitotoxicity by conjugated linoleic acid. J. Neurochem. 2010, 115, 123–130. [Google Scholar] [CrossRef]
- Weigel, T.K.; Kulas, J.A.; Ferris, H.A. Oxidized cholesterol species as signaling molecules in the brain: Diabetes and Alzheimer’s disease. Neuronal Signal. 2019, 3, NS20190068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henn, R.E.; Elzinga, S.E.; Glass, E.; Parent, R.; Guo, K.; Allouch, A.A.; Mendelson, F.E.; Hayes, J.; Webber-Davis, I.; Murphy, G.G.; et al. Obesity-induced neuroinflammation and cognitive impairment in young adult versus middle-aged mice. Immun. Ageing 2022, 19, 67. [Google Scholar] [CrossRef]
- Kanoke, A.; Nishijima, Y.; Ljungberg, M.; Omodaka, S.; Yang, S.Y.; Wong, S.; Rabiller, G.; Tominaga, T.; Hsieh, C.L.; Liu, J. The effect of type 2 diabetes on CD36 expression and the uptake of oxLDL: Diabetes affects CD36 and oxLDL uptake. Exp. Neurol. 2020, 334, 113461. [Google Scholar] [CrossRef]
- Kothari, V.; Luo, Y.; Tornabene, T.; O’Neill, A.M.; Greene, M.W.; Geetha, T.; Babu, J.R. High fat diet induces brain insulin resistance and cognitive impairment in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 499–508. [Google Scholar] [CrossRef]
- Avgerinos, K.I.; Mullins, R.J.; Vreones, M.; Mustapic, M.; Chen, Q.; Melvin, D.; Kapogiannis, D.; Egan, J.M. Empagliflozin Induced Ketosis, Upregulated IGF-1/Insulin Receptors and the Canonical Insulin Signaling Pathway in Neurons, and Decreased the Excitatory Neurotransmitter Glutamate in the Brain of Non-Diabetics. Cells 2022, 11, 3372. [Google Scholar] [CrossRef] [PubMed]
- Attia, H.; Albuhayri, S.; Alaraidh, S.; Alotaibi, A.; Yacoub, H.; Mohamad, R.; Al-Amin, M. Biotin, coenzyme Q10, and their combination ameliorate aluminium chloride-induced Alzheimer’s disease via attenuating neuroinflammation and improving brain insulin signaling. J. Biochem. Mol. Toxicol. 2020, 34, e22519. [Google Scholar] [CrossRef] [PubMed]
- Kuga, G.K.; Munoz, V.R.; Gaspar, R.C.; Nakandakari, S.; da Silva, A.S.R.; Botezelli, J.D.; Leme, J.; Gomes, R.J.; de Moura, L.P.; Cintra, D.E.; et al. Impaired insulin signaling and spatial learning in middle-aged rats: The role of PTP1B. Exp. Gerontol. 2018, 104, 66–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lalert, L.; Ji-Au, W.; Srikam, S.; Chotipinit, T.; Sanguanrungsirikul, S.; Srikiatkhachorn, A.; Maneesri-le Grand, S. Alterations in Synaptic Plasticity and Oxidative Stress Following Long-Term Paracetamol Treatment in Rat Brain. Neurotox. Res. 2020, 37, 455–468. [Google Scholar] [CrossRef]
- Callens, M.; Kraskovskaya, N.; Derevtsova, K.; Annaert, W.; Bultynck, G.; Bezprozvanny, I.; Vervliet, T. The role of Bcl-2 proteins in modulating neuronal Ca(2+) signaling in health and in Alzheimer’s disease. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118997. [Google Scholar] [CrossRef]
- Zhang, R.; Xue, Y.Y.; Lu, S.D.; Wang, Y.; Zhang, L.M.; Huang, Y.L.; Signore, A.P.; Chen, J.; Sun, F.Y. Bcl-2 enhances neurogenesis and inhibits apoptosis of newborn neurons in adult rat brain following a transient middle cerebral artery occlusion. Neurobiol. Dis. 2006, 24, 345–356. [Google Scholar] [CrossRef]
- Eugenin, J.; Vecchiola, A.; Murgas, P.; Arroyo, P.; Cornejo, F.; von Bernhardi, R. Expression Pattern of Scavenger Receptors and Amyloid-beta Phagocytosis of Astrocytes and Microglia in Culture are Modified by Acidosis: Implications for Alzheimer’s Disease. J. Alzheimers Dis. 2016, 53, 857–873. [Google Scholar] [CrossRef]
- Husemann, J.; Loike, J.D.; Anankov, R.; Febbraio, M.; Silverstein, S.C. Scavenger receptors in neurobiology and neuropathology: Their role on microglia and other cells of the nervous system. Glia 2002, 40, 195–205. [Google Scholar] [CrossRef]
- Luo, R.; Su, L.Y.; Li, G.; Yang, J.; Liu, Q.; Yang, L.X.; Zhang, D.F.; Zhou, H.; Xu, M.; Fan, Y.; et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy 2020, 16, 52–69. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Chiang, M.C.; Huang, Y.C.; Yeh, T.H.; Shih, H.Y.; Liu, H.F.; Chen, H.Y.; Wang, C.P.; Cheng, Y.C. Pparalpha deficiency inhibits the proliferation of neuronal and glial precursors in the zebrafish central nervous system. Dev. Dyn. 2018, 247, 1264–1275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mou, C.; Liu, B.; Wang, M.; Jiang, M.; Han, T. PGC-1-related coactivator (PRC) is an important regulator of microglia M2 polarization. J. Mol. Neurosci. 2015, 55, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Rueda, D.; Guerra-Ojeda, S.; Aldasoro, M.; Iradi, A.; Obrador, E.; Ortega, A.; Mauricio, M.D.; Vila, J.M.; Valles, S.L. Astrocytes protect neurons from Abeta1-42 peptide-induced neurotoxicity increasing TFAM and PGC-1 and decreasing PPAR-gamma and SIRT-1. Int. J. Med. Sci. 2015, 12, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.H.; Sinha, S.R.; Tanaka, K.; Rothstein, J.D.; Bergles, D.E. Astrocyte glutamate transporters regulate metabotropic glutamate receptor-mediated excitation of hippocampal interneurons. J. Neurosci. 2004, 24, 4551–4559. [Google Scholar] [CrossRef] [Green Version]
- Clyburn, C.; Browning, K.N. Role of astroglia in diet-induced central neuroplasticity. J. Neurophysiol. 2019, 121, 1195–1206. [Google Scholar] [CrossRef]
- Nakayama, T.; Mikoshiba, K.; Akagawa, K. The cell- and tissue-specific transcription mechanism of the TATA-less syntaxin 1A gene. FASEB J. 2016, 30, 525–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo, J.; Florit, S.; Giralt, M.; Ferrer, B.; Keller, C.; Pilegaard, H. Transgenic mice with astrocyte-targeted production of interleukin-6 are resistant to high-fat diet-induced increases in body weight and body fat. Brain. Behav. Immun. 2010, 24, 119–126. [Google Scholar] [CrossRef]
- Quintana, A.; Erta, M.; Ferrer, B.; Comes, G.; Giralt, M.; Hidalgo, J. Astrocyte-specific deficiency of interleukin-6 and its receptor reveal specific roles in survival, body weight and behavior. Brain. Behav. Immun. 2013, 27, 162–173. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jo, D.; Yoon, G.; Lim, Y.; Kim, Y.; Song, J. Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions. Int. J. Mol. Sci. 2023, 24, 6235. https://doi.org/10.3390/ijms24076235
Jo D, Yoon G, Lim Y, Kim Y, Song J. Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions. International Journal of Molecular Sciences. 2023; 24(7):6235. https://doi.org/10.3390/ijms24076235
Chicago/Turabian StyleJo, Danbi, Gwangho Yoon, Yeonghwan Lim, Youngkook Kim, and Juhyun Song. 2023. "Profiling and Cellular Analyses of Obesity-Related circRNAs in Neurons and Glia under Obesity-like In Vitro Conditions" International Journal of Molecular Sciences 24, no. 7: 6235. https://doi.org/10.3390/ijms24076235




