Stool Glycoproteomics Signatures of Pre-Cancerous Lesions and Colorectal Cancer
Abstract
1. Introduction
2. Results
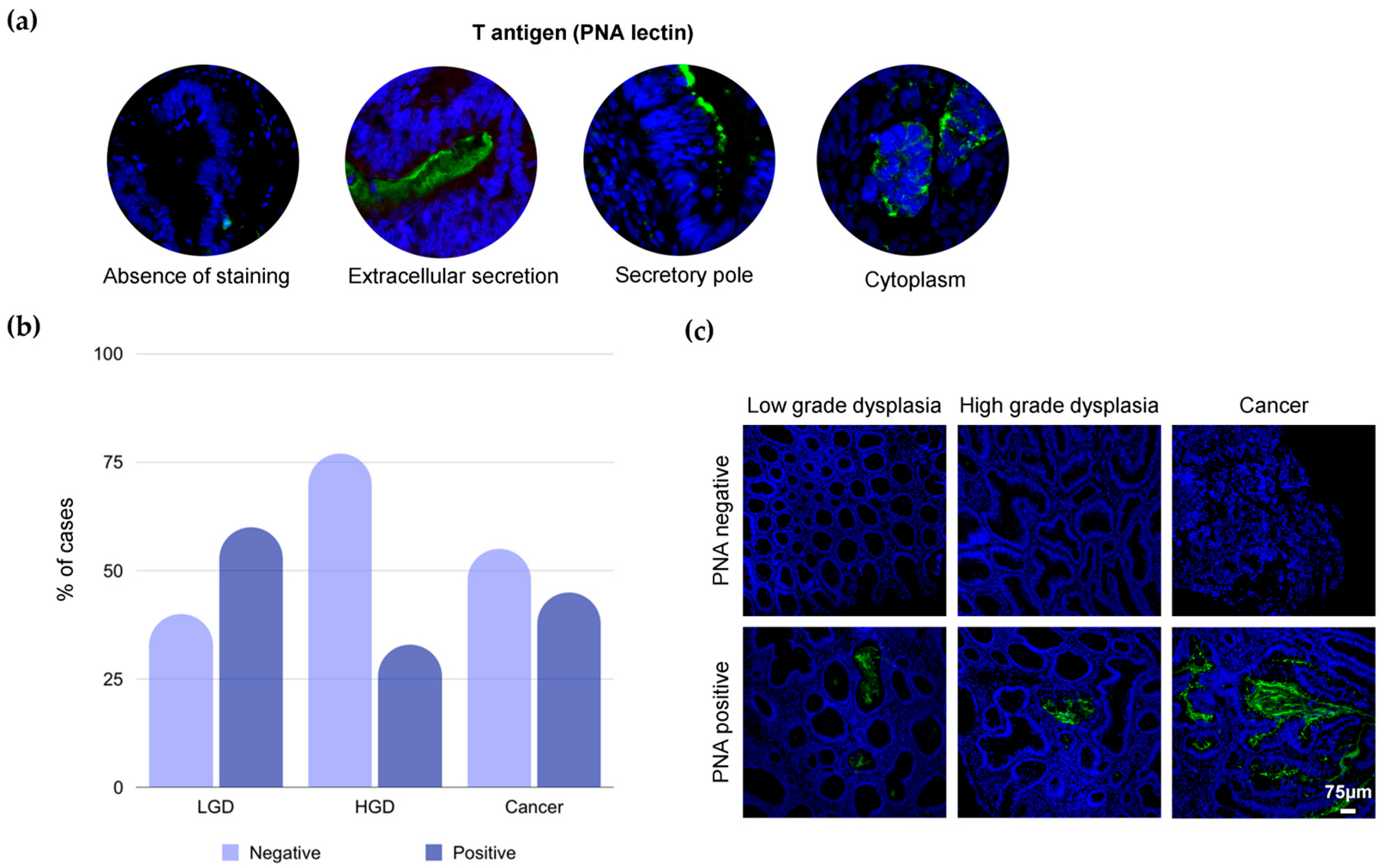
2.1. T-Antigen in Colorectal Tissues
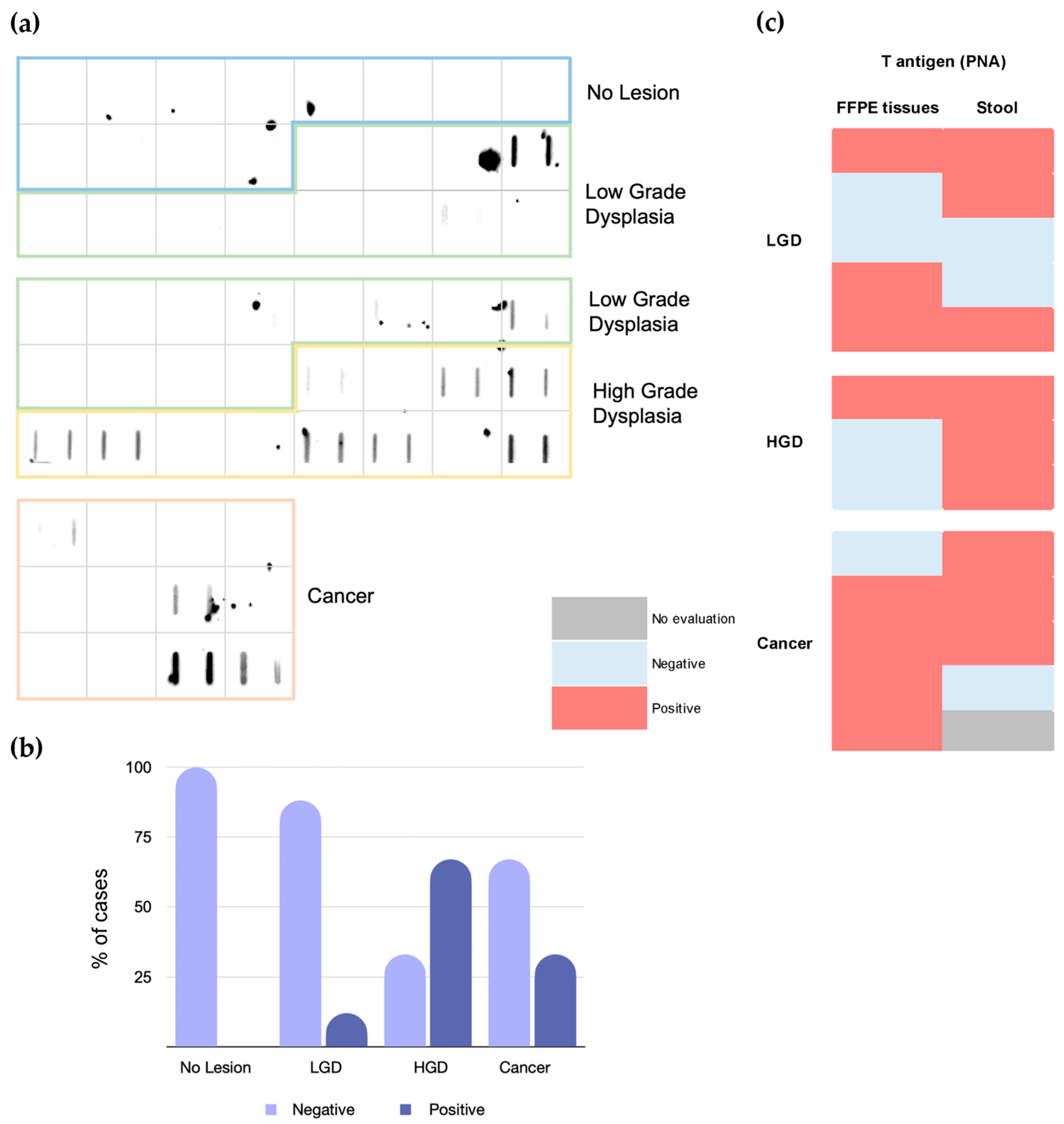
2.2. T-Antigen in Stool
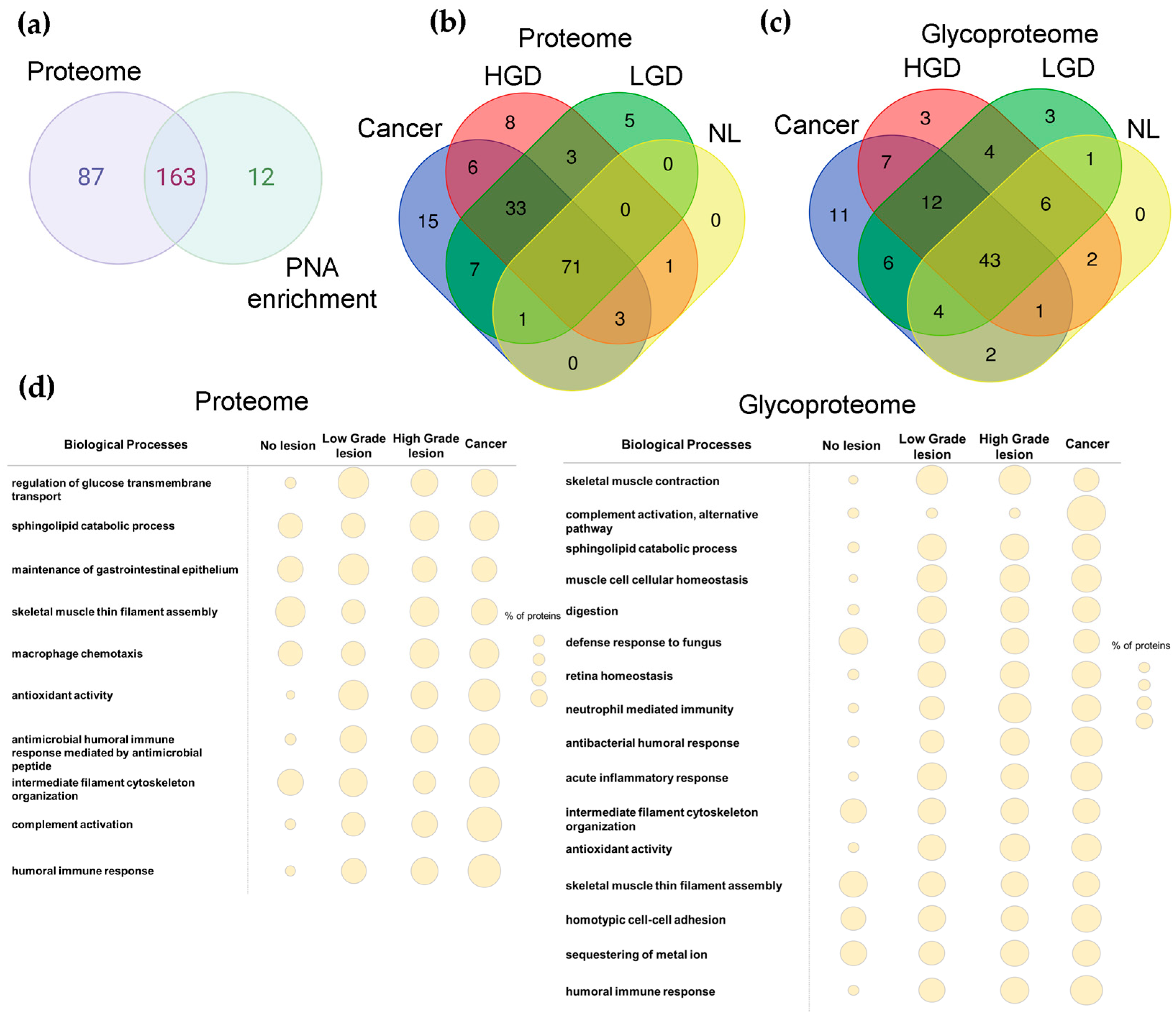
2.3. Proteomics and Glycoproteomics
3. Discussion
4. Materials and Methods
4.1. FFPE Tissues for Immunohistochemistry Studies
4.2. Patient Stool Sample Set
4.3. Immunofluorescence for the T-Antigen
4.4. Protein Extraction
4.5. Lectin Blotting
4.6. (Glyco)Proteomics
4.7. Bioinformatics
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schreuders, E.H.; Ruco, A.; Rabeneck, L.; Schoen, R.E.; Sung, J.J.; Young, G.P.; Kuipers, E.J. Colorectal cancer screening: A global overview of existing programmes. Gut 2015, 64, 1637–1649. [Google Scholar] [CrossRef] [PubMed]
- Simon, K. Colorectal cancer development and advances in screening. Clin. Interv. Aging. 2016, 11, 967–976. [Google Scholar] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Young, G.P.; Rabeneck, L.; Winawer, S.J. The Global Paradigm Shift in Screening for Colorectal Cancer. Gastroenterology 2019, 156, 843–851.e2. [Google Scholar] [CrossRef] [PubMed]
- Stracci, F.; Zorzi, M.; Grazzini, G. Colorectal cancer screening: Tests, strategies, and perspectives. Front. Public Health 2014, 2, 210. [Google Scholar] [CrossRef]
- Robertson, D.J.; Lee, J.K.; Boland, C.R.; Dominitz, J.A.; Giardiello, F.M.; Johnson, D.A.; Kaltenbach, T.; Lieberman, D.; Levin, T.R.; Rex, D.K. Recommendations on Fecal Immunochemical Testing to Screen for Colorectal Neoplasia: A Consensus Statement by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2017, 152, 1217–1237.e3. [Google Scholar] [CrossRef] [PubMed]
- Beniwal, S.S.; Lamo, P.; Kaushik, A.; Lorenzo-Villegas, D.L.; Liu, Y.; MohanaSundaram, A. Current Status and Emerging Trends in Colorectal Cancer Screening and Diagnostics. Biosensors 2023, 13, 926. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, T.; Kato, J.; Yamaji, Y.; Wada, R.; Mitsushima, T.; Shiratori, Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology 2005, 129, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Areia, M.; Fuccio, L.; Hassan, C.; Dekker, E.; Dias-Pereira, A.; Dinis-Ribeiro, M. Cost-utility analysis of colonoscopy or faecal immunochemical test for population-based organised colorectal cancer screening. United Eur. Gastroenterol. J. 2019, 7, 105–113. [Google Scholar] [CrossRef]
- Saftoiu, A.; Hassan, C.; Areia, M.; Bhutani, M.S.; Bisschops, R.; Bories, E.; Cazacu, I.M.; Dekker, E.; Deprez, P.H.; Pereira, S.P.; et al. Role of gastrointestinal endoscopy in the screening of digestive tract cancers in Europe: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy 2020, 52, 293–304. [Google Scholar] [CrossRef]
- Das, V.; Kalita, J.; Pal, M. Predictive and prognostic biomarkers in colorectal cancer: A systematic review of recent advances and challenges. Biomed. Pharmacother. 2017, 87, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Wuhrer, M.; Rombouts, Y. Glycosylation characteristics of colorectal cancer. Adv. Cancer Res. 2015, 126, 203–256. [Google Scholar] [PubMed]
- Madunic, K.; Mayboroda, O.A.; Zhang, T.; Weber, J.; Boons, G.J.; Morreau, H.; van Vlierberghe, R.; van Wezel, T.; Lageveen-Kammeijer, G.S.M.; Wuhrer, M. Specific (sialyl-)Lewis core 2 O-glycans differentiate colorectal cancer from healthy colon epithelium. Theranostics 2022, 12, 4498–4512. [Google Scholar] [CrossRef] [PubMed]
- Madunic, K.; Zhang, T.; Mayboroda, O.A.; Holst, S.; Stavenhagen, K.; Jin, C.; Karlsson, N.G.; Lageveen-Kammeijer, G.S.M.; Wuhrer, M. Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell Mol. Life Sci. 2021, 78, 337–350. [Google Scholar] [CrossRef]
- Cao, Y.; Stosiek, P.; Springer, G.F.; Karsten, U. Thomsen-Friedenreich-related carbohydrate antigens in normal adult human tissues: A systematic and comparative study. Histochem. Cell Biol. 1996, 106, 197–207. [Google Scholar] [CrossRef]
- Cao, Y.; Karsten, U.R.; Liebrich, W.; Haensch, W.; Springer, G.F.; Schlag, P.M. Expression of Thomsen-Friedenreich-related antigens in primary and metastatic colorectal carcinomas. A reevaluation. Cancer 1995, 76, 1700–1708. [Google Scholar] [CrossRef]
- Li, F.; Glinskii, O.V.; Mooney, B.P.; Rittenhouse-Olson, K.; Pienta, K.J.; Glinsky, V.V. Cell surface Thomsen-Friedenreich proteome profiling of metastatic prostate cancer cells reveals potential link with cancer stem cell-like phenotype. Oncotarget 2017, 8, 98598–98608. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Polom, K.; Williams, C.; Polonia, A.; Guergova-Kuras, M.; Karlsson, N.G.; Roviello, F.; Magalhães, A.; Reis, C.A. The Thomsen-Friedenreich Antigen: A Highly Sensitive and Specific Predictor of Microsatellite Instability in Gastric Cancer. J. Clin. Med. 2018, 7, 256. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, N.; Bane, S.M.; Ahire, S.D.; Ingle, A.D.; Kalraiya, R.D. Poly N-acetyllactosamine substitutions on N- and not O-oligosaccharides or Thomsen-Friedenreich antigen facilitate lung specific metastasis of melanoma cells via galectin-3. Glycoconj. J. 2009, 26, 445–456. [Google Scholar] [CrossRef]
- Takanami, I. Expression of Thomsen-Friedenreich antigen as a marker of poor prognosis in pulmonary adenocarcinoma. Oncol. Rep. 1999, 6, 341–344. [Google Scholar] [CrossRef]
- Santos-Silva, F.; Fonseca, A.; Caffrey, T.; Carvalho, F.; Mesquita, P.; Reis, C.; Almeida, R.; David, L.; Hollingsworth, M.A. Thomsen-Friedenreich antigen expression in gastric carcinomas is associated with MUC1 mucin VNTR polymorphism. Glycobiology 2005, 15, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Trabbic, K.R.; Whalen, K.; Abarca-Heideman, K.; Xia, L.; Temme, J.S.; Edmondson, E.F.; Gildersleeve, J.C.; Barchi, J.J., Jr. A Tumor-Selective Monoclonal Antibody from Immunization with a Tumor-Associated Mucin Glycopeptide. Sci. Rep. 2019, 9, 5662. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Ferreira, D.; Azevedo, R.; Freitas, R.; Fernandes, E.; Relvas-Santos, M.; Gaiteiro, C.; Soares, J.; Cotton, S.; Teixeira, B.; et al. Glycoproteomics identifies HOMER3 as a potentially targetable biomarker triggered by hypoxia and glucose deprivation in bladder cancer. J. Exp. Clin. Cancer Res. 2021, 40, 191. [Google Scholar] [CrossRef] [PubMed]
- Bojar, D.; Meche, L.; Meng, G.; Eng, W.; Smith, D.F.; Cummings, R.D.; Mahal, L.K. A Useful Guide to Lectin Binding: Machine-Learning Directed Annotation of 57 Unique Lectin Specificities. ACS Chem. Biol. 2022, 17, 2993–3012. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, H.; Yang, H.; Zhou, J.; Zhao, L.; Zhang, F. Glucose metabolism and glycosylation link the gut microbiota to autoimmune diseases. Front. Immunol. 2022, 13, 952398. [Google Scholar] [CrossRef] [PubMed]
- Leao, B.; Wen, X.; Duarte, H.O.; Gullo, I.; Goncalves, G.; Pontes, P.; Castelli, C.; Diniz, F.; Mereiter, S.; Gomes, J.; et al. Expression of Thomsen-Friedenreich Antigen in Colorectal Cancer and Association with Microsatellite Instability. Int. J. Mol. Sci. 2021, 22, 1340. [Google Scholar] [CrossRef] [PubMed]
- Cayrefourcq, L.; Thomas, F.; Mazard, T.; Assenat, E.; Assou, S.; Alix-Panabieres, C. Selective treatment pressure in colon cancer drives the molecular profile of resistant circulating tumor cell clones. Mol. Cancer 2021, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.S.; Yamashita, Y.; Kato, Y.; Nakata, B.; Sawada, T.; Sowa, M. Prognostic significance of T antigen expression in patients with gastric carcinoma. Cancer 1996, 77, 1768–1773. [Google Scholar] [CrossRef]
- Springer, G.F.; Desai, P.R.; Murthy, M.S.; Scanlon, E.F. Human carcinoma-associated precursor antigens of the NM blood group system. J. Surg. Oncol. 1979, 11, 95–106. [Google Scholar] [CrossRef]
- Yu, L.G.; Andrews, N.; Zhao, Q.; McKean, D.; Williams, J.F.; Connor, L.J.; Gerasimenko, O.V.; Hilkens, J.; Hirabayashi, J.; Kasai, K.; et al. Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J. Biol. Chem. 2007, 282, 773–781. [Google Scholar] [CrossRef]
- Khaldoyanidi, S.K.; Glinsky, V.V.; Sikora, L.; Glinskii, A.B.; Mossine, V.V.; Quinn, T.P.; Glinsky, G.V.; Sriramarao, P. MDA-MB-435 human breast carcinoma cell homo- and heterotypic adhesion under flow conditions is mediated in part by Thomsen-Friedenreich antigen-galectin-3 interactions. J. Biol. Chem. 2003, 278, 4127–4134. [Google Scholar] [CrossRef] [PubMed]
- Cotton, S.; Azevedo, R.; Gaiteiro, C.; Ferreira, D.; Lima, L.; Peixoto, A.; Fernandes, E.; Neves, M.; Neves, D.; Amaro, T.; et al. Targeted O-glycoproteomics explored increased sialylation and identified MUC16 as a poor prognosis biomarker in advanced-stage bladder tumours. Mol. Oncol. 2017, 11, 895–912. [Google Scholar] [CrossRef] [PubMed]
- Juge, N.; Tailford, L.; Owen, C.D. Sialidases from gut bacteria: A mini-review. Biochem. Soc. Trans. 2016, 44, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Tripathy, A.S. Alternative pathway of complement activation has a beneficial role against Chandipura virus infection. Med. Microbiol. Immunol. 2020, 209, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.J.; Plichta, D.R.; Asher, S.; Delsignore, M.; Jeong, T.; McGoldrick, J.; Staller, K.; Khalili, H.; Xavier, R.J.; Chung, D.C. Association of distinct microbial signatures with premalignant colorectal adenomas. Cell Host Microbe 2023, 31, 827–838.e3. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef] [PubMed]
- Radak, M.; Fallahi, H. Unraveling molecular similarities between colorectal polyps and colorectal cancer: A systems biology approach. Intest. Res. 2024. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.C.; Yu, J. Gut microbiota in colorectal cancer development and therapy. Nat. Rev. Clin. Oncol. 2023, 20, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Darebna, P.; Novak, P.; Kucera, R.; Topolcan, O.; Sanda, M.; Goldman, R.; Pompach, P. Changes in the expression of N- and O-glycopeptides in patients with colorectal cancer and hepatocellular carcinoma quantified by full-MS scan FT-ICR and multiple reaction monitoring. J. Proteom. 2017, 153, 44–52. [Google Scholar] [CrossRef]
- Ivancic, M.M.; Megna, B.W.; Sverchkov, Y.; Craven, M.; Reichelderfer, M.; Pickhardt, P.J.; Sussman, M.R.; Kennedy, G.D. Noninvasive Detection of Colorectal Carcinomas Using Serum Protein Biomarkers. J. Surg. Res. 2020, 246, 160–169. [Google Scholar] [CrossRef]
- Fiorito, V.; Tolosano, E. Hemopexin and Cancer. Int. J. Mol. Sci. 2022, 23, 160–169. [Google Scholar] [CrossRef]
- Bosch, L.J.W.; de Wit, M.; Pham, T.V.; Coupe, V.M.H.; Hiemstra, A.C.; Piersma, S.R.; Oudgenoeg, G.; Scheffer, G.L.; Mongera, S.; Sive Droste, J.T.; et al. Novel Stool-Based Protein Biomarkers for Improved Colorectal Cancer Screening: A Case-Control Study. Ann. Intern. Med. 2017, 167, 855–866. [Google Scholar] [CrossRef]
- Komor, M.A.; Bosch, L.J.; Coupe, V.M.; Rausch, C.; Pham, T.V.; Piersma, S.R.; Mongera, S.; Mulder, C.J.J.; Dekker, E.; Kuipers, E.J.; et al. Proteins in stool as biomarkers for non-invasive detection of colorectal adenomas with high risk of progression. J. Pathol. 2020, 250, 288–298. [Google Scholar] [CrossRef]
- Canesin, G.; Di Ruscio, A.; Li, M.; Ummarino, S.; Hedblom, A.; Choudhury, R.; Krzyzanowska, A.; Csizmadia, E.; Palominos, M.; Stiehm, A.; et al. Scavenging of Labile Heme by Hemopexin Is a Key Checkpoint in Cancer Growth and Metastases. Cell Rep. 2020, 32, 108181. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Takadate, T.; Mizuma, M.; Shima, H.; Suzuki, T.; Tachibana, T.; Shimura, M.; Hata, T.; Iseki, M.; Kawaguchi, K.; et al. Stromal expression of hemopexin is associated with lymph-node metastasis in pancreatic ductal adenocarcinoma. PLoS ONE 2020, 15, e0235904. [Google Scholar] [CrossRef]
- Miyamoto, S.; Ruhaak, L.R.; Stroble, C.; Salemi, M.R.; Phinney, B.; Lebrilla, C.B.; Leiserowitz, G.S. Glycoproteomic Analysis of Malignant Ovarian Cancer Ascites Fluid Identifies Unusual Glycopeptides. J. Proteome Res. 2016, 15, 3358–3376. [Google Scholar] [CrossRef] [PubMed]
- Chantaraamporn, J.; Champattanachai, V.; Khongmanee, A.; Verathamjamras, C.; Prasongsook, N.; Mingkwan, K.; Luevisadpibul, V.; Chutipongtanate, S.; Svasti, J. Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer. Proteomes 2020, 8, 26. [Google Scholar] [CrossRef]
- Bao, D.; Zhang, C.; Li, L.; Wang, H.; Li, Q.; Ni, L.; Lin, Y.; Huang, R.; Yang, Z.; Zhang, Y.; et al. Integrative Analysis of Complement System to Prognosis and Immune Infiltrating in Colon Cancer and Gastric Cancer. Front. Oncol. 2020, 10, 553297. [Google Scholar] [CrossRef] [PubMed]
- Bohana-Kashtan, O.; Pinna, L.A.; Fishelson, Z. Extracellular phosphorylation of C9 by protein kinase CK2 regulates complement-mediated lysis. Eur. J. Immunol. 2005, 35, 1939–1948. [Google Scholar] [CrossRef]
- Li, L.; Yang, H.; Li, Y.; Li, X.D.; Zeng, T.T.; Lin, S.X.; Zhu, Y.H.; Guan, X.Y. Hypoxia restrains the expression of complement component 9 in tumor-associated macrophages promoting non-small cell lung cancer progression. Cell Death Discov. 2018, 4, 63. [Google Scholar] [CrossRef]
- Cao, Z.Q.; Guo, X.L. The role of galectin-4 in physiology and diseases. Protein Cell. 2016, 7, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, E.M.; Hansen, G.H. Lipid raft organization and function in brush borders of epithelial cells. Mol. Membr. Biol. 2006, 23, 71–79. [Google Scholar] [CrossRef]
- Barrow, H.; Rhodes, J.M.; Yu, L.G. Simultaneous determination of serum galectin-3 and -4 levels detects metastases in colorectal cancer patients. Cell Oncol. 2013, 36, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Barrow, H.; Guo, X.; Wandall, H.H.; Pedersen, J.W.; Fu, B.; Zhao, Q.; Chen, C.; Rhodes, J.M.; Yu, L.G. Serum galectin-2, -4, and -8 are greatly increased in colon and breast cancer patients and promote cancer cell adhesion to blood vascular endothelium. Clin. Cancer Res. 2011, 17, 7035–7046. [Google Scholar] [CrossRef]
- Kume, H.; Muraoka, S.; Kuga, T.; Adachi, J.; Narumi, R.; Watanabe, S.; Kuwano, M.; Kodera, Y.; Matsushita, K.; Fukuoka, J.; et al. Discovery of colorectal cancer biomarker candidates by membrane proteomic analysis and subsequent verification using selected reaction monitoring (SRM) and tissue microarray (TMA) analysis. Mol. Cell Proteom. 2014, 13, 1471–1484. [Google Scholar] [CrossRef]
- Furuya, K.; Nakajima, M.; Tsunedomi, R.; Nakagami, Y.; Xu, M.; Matsui, H.; Tokumitsu, Y.; Shindo, Y.; Watanabe, Y.; Tomochika, S.; et al. High serum proteinase-3 levels predict poor progression-free survival and lower efficacy of bevacizumab in metastatic colorectal cancer. BMC Cancer 2024, 24, 165. [Google Scholar] [CrossRef]
- Kalimutho, M.; Blanco Gdel, V.; Gravina, P.; Cretella, M.; Mannucci, L.; Mannisi, E.; Formosa, A.; Pallone, F.; Federici, G.; Bernardini, S. Quantitative denaturing high performance liquid chromatography (Q-dHPLC) detection of APC long DNA in faeces from patients with colorectal cancer. Clin. Chem. Lab. Med. 2010, 48, 1303–1311. [Google Scholar] [CrossRef]
- Phua, L.C.; Chue, X.P.; Koh, P.K.; Cheah, P.Y.; Chan, E.C.; Ho, H.K. Global fecal microRNA profiling in the identification of biomarkers for colorectal cancer screening among asians. Oncol. Rep. 2014, 32, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Gaiteiro, C.; Soares, J.; Relvas-Santos, M.; Peixoto, A.; Ferreira, D.; Paulo, P.; Brandão, A.; Fernandes, E.; Azevedo, R.; Palmeira, C.; et al. Glycoproteogenomics characterizes the CD44 splicing code associated with bladder cancer invasion. Theranostics 2022, 12, 3150–3177. [Google Scholar] [CrossRef]
- Peixoto, A.; Ferreira, D.; Miranda, A.; Relvas-Santos, M.; Freitas, R.; Veth, T.S.; Brandão, A.; Ferreira, E.; Paulo, P.; Cardoso, M.; et al. Multilevel Plasticity and Altered Glycosylation Drive Aggressiveness in Hypoxic and Glucose-Deprived Bladder Cancer Cells. bioRxiv 2023. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

| T-Antigen Positive | T-Antigen Negative | ||
|---|---|---|---|
| n (%) | n (%) | p-Value | |
| Stage | |||
| I | 1 (5.55) | 0 (0) | |
| II | 0 (0) | 4 (18.18) | |
| III | 3 (16.67) | 7 (31.82) | 0.085 |
| IV | 14 (77.78) | 11 (50.00) | |
| Tumor (T) | |||
| T1 | 0 (0) | 1 (4.76) | |
| T2 | 1 (5.55) | 0 (0) | |
| T3 | 14 (77.78) | 17 (80.95) | 0.558 |
| T4 | 3 (16.67) | 3 (14.29) | |
| Missing information | 1 (4.76) | ||
| Lymph node metastases (N) | |||
| N0 | 5 (27.78) | 6 (27.27) | |
| N1 | 7 (38.89) | 5 (22.73) | |
| N2 | 5 (27.78) | 9 (40.91) | 0.679 |
| N3 | 1 (5.55) | 2 (9.09) | |
| Distant metastases (M) | |||
| M0 | 4 (22.22) | 12 (54.5) | 0.039 |
| M1 | 14 (77.78) | 10 (45.5) | |
| Tumor location | |||
| Right colon | 3 (16.67) | 4 (18.19) | |
| Left colon | 13 (72.22) | 10 (45.45) | 0.152 |
| Rectum | 2 (11.11) | 8 (36.36) |
| N (%) | |
|---|---|
| Stage | |
| I | 1 (2.5) |
| II | 4 (10) |
| III | 10 (25) |
| IV | 25 (65) |
| Tumor (T) | |
| T1 | 1 (2.5) |
| T2 | 1 (2.5) |
| T3 | 31 (77.5) |
| T4 | 6 (15) |
| Missing Information | 1 (2.5) |
| Lymph node metastases (N) | |
| N0 | 11 (27.5) |
| N1 | 12 (30) |
| N2 | 14 (35) |
| N3 | 3 (7.5) |
| Tumor Location | |
| Right colon | 7 (17.5) |
| Left colon | 23 (57.5) |
| Rectum | 10 (25) |
| Characteristics | No Lesion (n = 12) | Low Grade Dysplasia (n = 24) | High-Grade Dysplasia (n = 12) | Cancer (n = 12) |
|---|---|---|---|---|
| Age (mean, min–max) | 58.0 (50–72) | 56.0 (49–74) | 63.5 (40–85) | 64.5 (51–89) |
| Gender | ||||
| Male | 37.5% | 37.5% | 58.3% | 54.2% |
| Female | 62.5% | 62.5% | 41.7% | 45.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, J.; Eiras, M.; Ferreira, D.; Santos, D.A.R.; Relvas-Santos, M.; Santos, B.; Gonçalves, M.; Ferreira, E.; Vieira, R.; Afonso, L.P.; et al. Stool Glycoproteomics Signatures of Pre-Cancerous Lesions and Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 3722. https://doi.org/10.3390/ijms25073722
Soares J, Eiras M, Ferreira D, Santos DAR, Relvas-Santos M, Santos B, Gonçalves M, Ferreira E, Vieira R, Afonso LP, et al. Stool Glycoproteomics Signatures of Pre-Cancerous Lesions and Colorectal Cancer. International Journal of Molecular Sciences. 2024; 25(7):3722. https://doi.org/10.3390/ijms25073722
Chicago/Turabian StyleSoares, Janine, Mariana Eiras, Dylan Ferreira, Daniela A. R. Santos, Marta Relvas-Santos, Beatriz Santos, Martina Gonçalves, Eduardo Ferreira, Renata Vieira, Luís Pedro Afonso, and et al. 2024. "Stool Glycoproteomics Signatures of Pre-Cancerous Lesions and Colorectal Cancer" International Journal of Molecular Sciences 25, no. 7: 3722. https://doi.org/10.3390/ijms25073722
APA StyleSoares, J., Eiras, M., Ferreira, D., Santos, D. A. R., Relvas-Santos, M., Santos, B., Gonçalves, M., Ferreira, E., Vieira, R., Afonso, L. P., Santos, L. L., Dinis-Ribeiro, M., Lima, L., & Ferreira, J. A. (2024). Stool Glycoproteomics Signatures of Pre-Cancerous Lesions and Colorectal Cancer. International Journal of Molecular Sciences, 25(7), 3722. https://doi.org/10.3390/ijms25073722








