Preliminary Analysis of Life within a Former Subglacial Lake Sediment in Antarctica
Abstract
:1. Introduction
2. Experimental Section
2.1. Sample Site
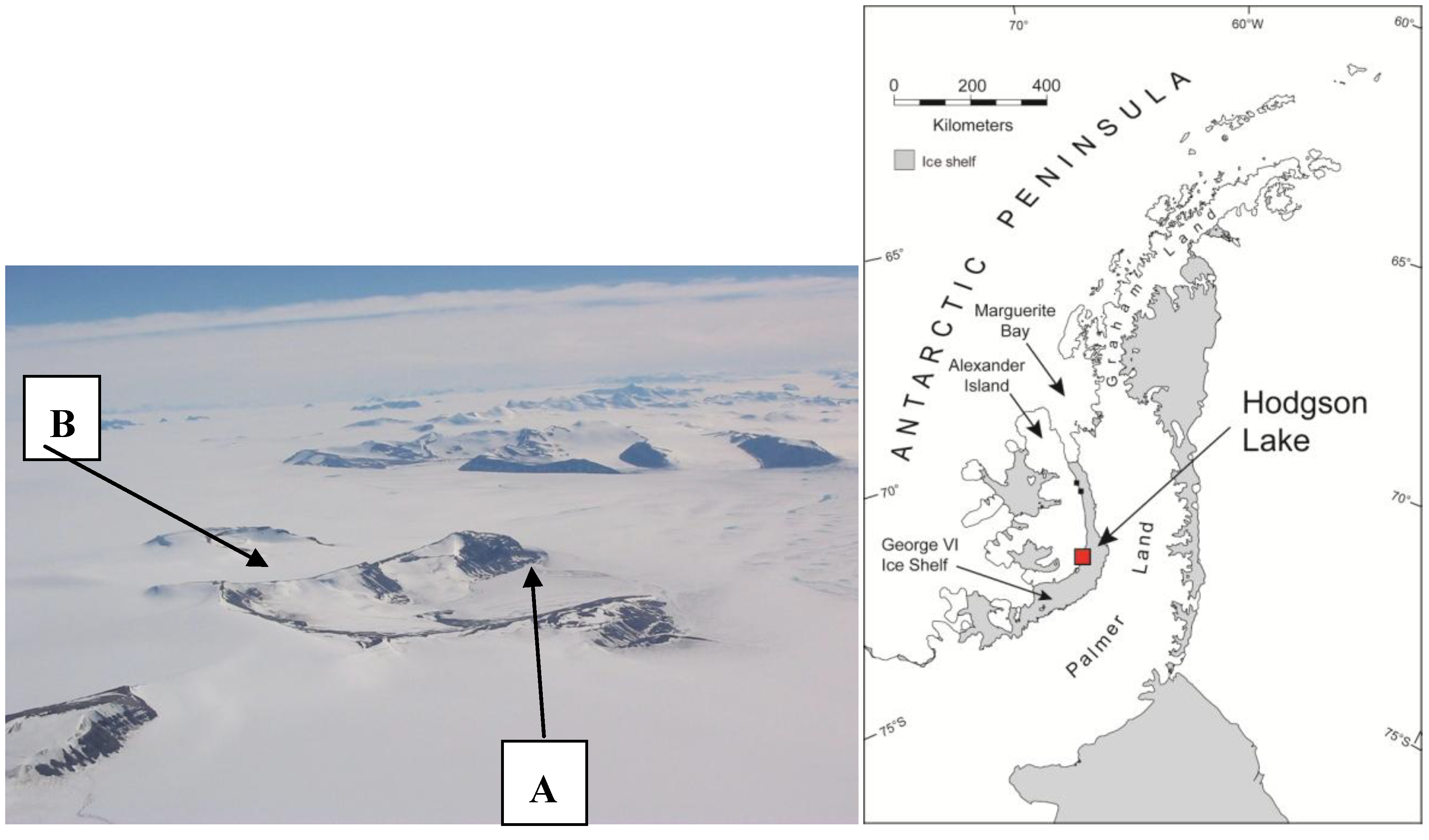
2.2. Sample Recovery
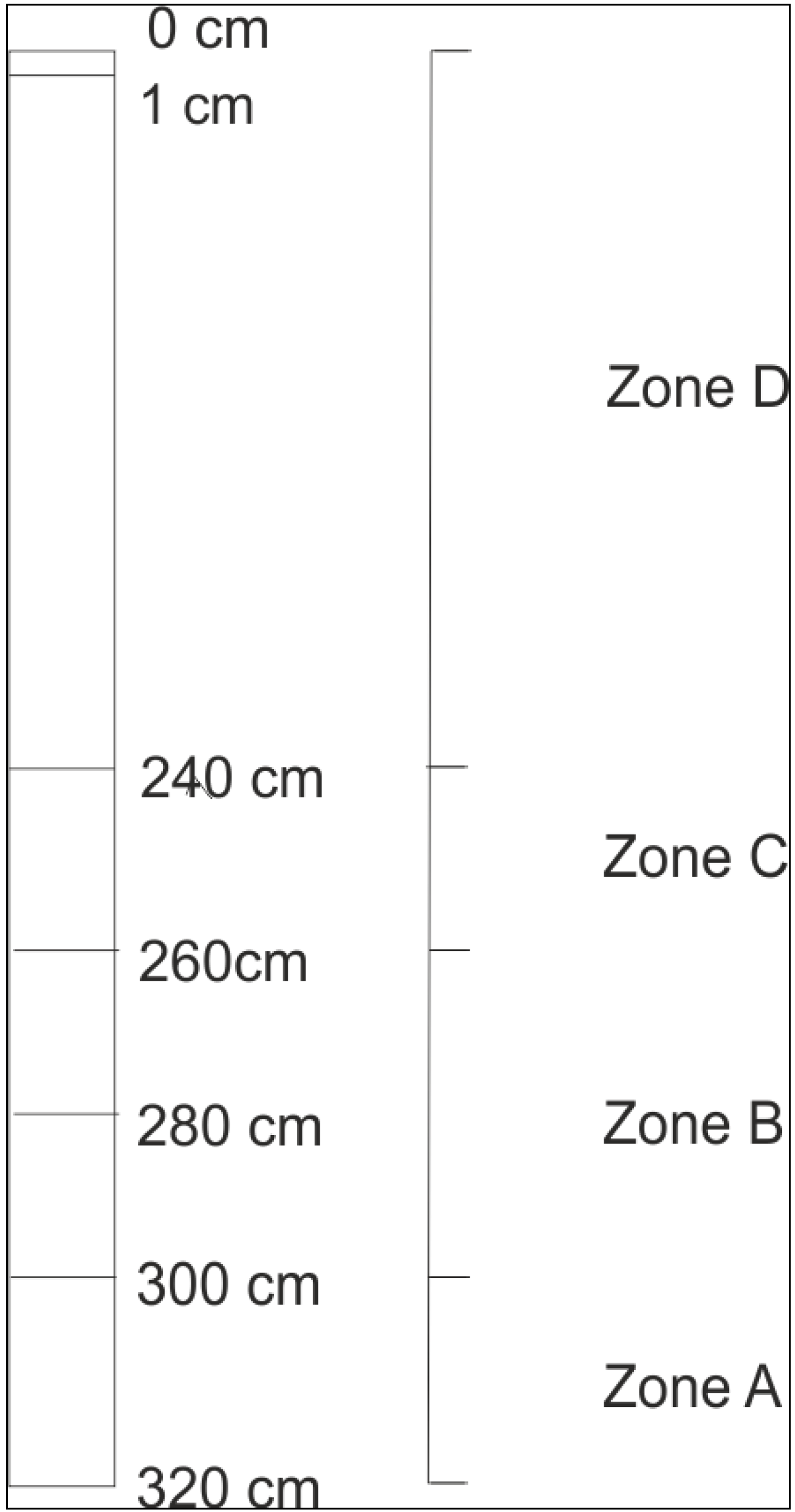
2.3. Microscopy (DAPI Staining)
2.4. Microscopy (Fluorescence in Situ Hybridization)
2.5. Microscopy (Scanning Electron Microscopy)
2.6. Direct Culture
2.7. DNA Extraction and PCR Amplification
2.8. 454 Pyrosequencing
2.9. Data Analysis
3. Results and Discussion
3.1. Microscopy
3.2. Direct Culture
3.3. 454 Pyrosequencing
3.4. SSU
3.5. RDP Analysis
| Organism | Abundance | Avg. eValue exponent | Avg. % ident. | Avg. align length |
|---|---|---|---|---|
| uncultured bacterium | 12424 | −71.72 | 99.46 | 139.44 |
| Pirellula staleyi | 9869 | −39.89 | 99.83 | 83.06 |
| uncultured delta proteobacterium | 6903 | −48.08 | 99.72 | 97.44 |
| unassigned | 5481 | −65.37 | 99.62 | 127.16 |
| Thermobaculum terrenum | 5083 | −43.62 | 99.64 | 89.4 |
| Rhodopirellula baltica | 3021 | −32.16 | 99.61 | 70.38 |
| Dehalococcoides ethenogenes | 2917 | −35.55 | 98.97 | 78.98 |
| unidentified | 2859 | −43.24 | 99.08 | 91.3 |
| Coptotermes formosanus | 2627 | −34.79 | 99.59 | 75.26 |
| uncultured Dehalococcoides sp. | 2494 | −31.23 | 100 | 67.85 |
| Spirochaeta aurantia | 2408 | −35.59 | 99.95 | 74.67 |
| Spirochaeta thermophila | 2401 | −37.59 | 100 | 78.31 |
| uncultured proteobacterium | 2356 | −52.95 | 99.91 | 104.68 |
| Methylococcus capsulatus | 2113 | −41.94 | 99.42 | 86.89 |
| Thermomicrobium roseum | 1429 | −33.81 | 99.6 | 73.24 |
| Actinosynnema mirum | 1394 | −62.16 | 94.94 | 135.81 |
| Mycobacterium leprae | 1336 | −56.61 | 95.66 | 123.56 |
| Streptomyces microflavus | 1333 | −38 | 100 | 79.33 |
| Protochlamydia naegleriophila | 1300 | −37.05 | 99.79 | 77.86 |
| Leptospirillum ferrodiazotrophum | 1179 | −36.33 | 99.74 | 77.45 |
| Conexibacter woesei | 1128 | −54.7 | 99.04 | 110.35 |
| Amycolatopsis sp. GY182 | 1076 | −34.33 | 100 | 72.67 |
| Nocardioides sp. AL050511-10 | 820 | −44.68 | 99.82 | 91.15 |
| Propionibacterium acnes | 800 | −74.66 | 99.64 | 143.79 |
| Frankia sp. | 785 | −53.45 | 99.48 | 107.5 |
| D. lykanthroporepellens | 616 | −29.24 | 99.97 | 64.3 |
| Rhodopirellula sp. SM49 | 512 | −76.38 | 99.08 | 149.25 |
| Thermoleophilum album | 509 | −54.25 | 99.7 | 107.06 |
| Microbacterium arborescens | 488 | −107 | 99.39 | 199.31 |
| Propionibacterium acidifaciens | 466 | −36.97 | 100 | 77.95 |
| Antarctic soil bacterium 2-1 | 448 | −114.1 | 97.93 | 219.8 |
| Kocuria rhizophila | 397 | −79.08 | 98.53 | 154.58 |
| Atopostipes suicloacalis | 386 | −36.67 | 99.23 | 78.67 |
| uncultured alpha proteobacterium | 379 | −62.04 | 99.7 | 120.96 |
| Thermodesulfovibrio islandicus | 366 | −32.29 | 100 | 69.32 |
| Sphaerobacter thermophilus | 347 | −29.59 | 99.76 | 65.23 |
| Lechevalieria aerocolonigenes | 333 | −48.14 | 98.9 | 102.14 |
| Lentzea violacea | 333 | −35.33 | 99.78 | 74.83 |
| Pedosphaera parvula | 328 | −36.62 | 100 | 77.12 |
| unidentified marine eubacterium | 316 | −41.24 | 99.01 | 87.93 |
| Simkania negevensis | 307 | −38.3 | 100 | 79.48 |
| Uncultured γ proteobacterium | 303 | −94.04 | 99.83 | 175.23 |
| bacterium culture clone N47 | 280 | −32.28 | 99.69 | 70.6 |
| uncultured soil bacterium | 279 | −57.41 | 99.57 | 113.29 |
| Candidatus Koribacter versatilis | 260 | −51 | 99.8 | 101.57 |
| Acidothermus cellulolyticus | 250 | −48.39 | 99.86 | 96.44 |
| Planctomyces limnophilus | 243 | −34.37 | 100 | 73.11 |
| Desulfothermus naphthae | 241 | −40.8 | 100 | 83.8 |
| Staphylococcus carnosus | 213 | −29 | 100 | 63 |
| Rhodococcus equi | 211 | −30.33 | 100 | 65.67 |
| Thermodesulfobium narugense | 211 | −36.18 | 97.78 | 80.77 |
3.6. Greengenes Analysis
4. Conclusions
| Culture | FISH | 454 |
|---|---|---|
| 3x Sporosarcina sp. | LGC354 no hybridization | Firmicutes 5.3% and 6th most abundant sequence type |
| 15x Arthrobacter sp. | HGC263 no hybridization | Actinobacteria 23.8% most abundant of sequences |
| 2x Streptomyces sp. | HGC263 no hybridization | Actinobacteria 23.8% most abundant of sequences |
| Present | EUB338 hybridization | Most sequences |
| No culture obtained | ARCH915 no hybridization | Archaea 0.5% of known sequences |
| No culture obtained | ALF968 hybridization | Alphaproteobacteria 1.2% of known sequences |
| No culture obtained | GAM42a hybridization | Gammaproteobacteria 5.6% of known sequences |
| No culture obtained | BET42a hybridization | Betaproteobacteria 0.3% of known sequences |
| No culture obtained | CF319 hybridization | Bacteroidetes 2.2% of known sequences |
| No culture obtained | SRB385 no hybridization | Deltaproteobacteria 10.8% of known sequences |
| No culture obtained | ANME-1-350 no hybridization | Some Archaeal methanogens detected |
| Sediment type | Position | Clone | |
|---|---|---|---|
| Shivaji et al. [62] | Freshwater lake | Upper (18–22 cm) | |
| Middle (60–64 cm) | |||
| Lower (100–104 cm) | |||
| Combined | Proteobacteria | ||
| Bacteroidetes | |||
| Actinobacteria | |||
| Firmicutes | |||
| Caldiserica | |||
| Sjöling & Cowan [63] | Glacial meltwater | Alphaproteobacteria | |
| lake | Gammaproteobacteria | ||
| Deltaproteobacteria | |||
| Cytophaga-Flavobacterium-Bacteroides | |||
| Spirochaetaceae | |||
| Actinobacteria | |||
| Crenarchaeota deep-branching | |||
| Group 1 Marine Archaea | |||
| Brambilla et al. [64] | Mat samples | Surface | Proteobacteria |
| Lake Fryxell | Actinobacteria | ||
| Clostridium/Bacillus | |||
| Cytophaga-Flavobacterium-Bacteroides | |||
| Flavobacterium hibernum | |||
| Janthiniobacterium lividum | |||
| Arthrobacter flavusaerobic | |||
| Clostridium estertheticum | |||
| Hawes & Sutherland [65] | Benthic mat | Cyanobacteria | |
| Li et al. [66] | Lake sediment core | 1–20 cm | Alphaproteobacteria |
| 21–46 cm | Gammaproteobacteria | ||
| 46–59 cm | Deltaproteobacteria | ||
| Cytophaga-Flavobacterium-Bacteroides | |||
| Gemmatimonadetes | |||
| Firmicutes | |||
| and Actinobacteria | |||
| Bowman et al. [67] | Continental shelf | Deltaproteobacteria | |
| sediments | Gammaproteobacteria | ||
| 709 to 964 m | Flavobacteria | ||
| Planctomycetales | |||
| Archaea | |||
| Stackebrandt et al. [68] | Anaerobic mat | Firmicutes | |
| Lake Fryxell | Proteobacteria | ||
| Bacteriodetes | |||
| Many novel species | |||
| Purdy et al. [69] | Freshwater lake | Methanosaeta concilii | |
| Heywood | Limited archaeal diversity | ||
| Purdy et al. [69] | Marine Shallow Bay | Methanolobus | |
| Methanococcoides | |||
| Methanogenium | |||
| Desulfotalea/Desulforhopalus | |||
| Desulfofaba | |||
| Desulfosarcina | |||
| Desulfobacter | |||
| Desulfuromonas cluster | |||
| Bowman et al. [70] | Meromictic lake | Low G + C Gram-positive | |
| marine salinity | Prochlorococcus Cyanobacteria | ||
| anoxic | Diatom chloroplasts | ||
| and a | Deltaproteobacteria | ||
| Marine basin | Chlamydiales | ||
| meromictic | Spirochaetales | ||
| coastal | Euryarchaeota | ||
| Chen et al. [71] | Lake and deep sea | Arthrobacter ardleyensis | |
| Wang et al. [72] | Freshwater lake | Flavobacterium saliperosum sp. nov | |
| Mancuso et al. [53] | Methanogenic | ||
| Antarctic Lake | |||
| Karr et al. [73] | Lake Fryxell | Two clusters of methanogens |
4.1. Microbial Species Diversity
4.2. Summary
Acknowledgments
Conflict of Interest
References
- Robin, G.D.Q.; Swithinbank, C.W.M.; Smith, M.B.E. Radio echo exploration of the Antarctic ice sheet. In International Symposium on Antarctic Glaciological Exploration (ISAGE); International Association of Scientific Hydrology: Hanover, NH, USA, 1970; Publication number 86; pp. 97–115. [Google Scholar]
- Oswald, G.K.A.; Robin, G.D.Q. Lakes beneath Antarctic ice sheet. Nature 1973, 245, 251–254. [Google Scholar] [CrossRef]
- Siegert, M.J.; Tranter, M.; Ellis-Evans, J.C.; Priscu, J.C.; Lyons, W.B. The hydrochemistry of lake Vostok and the potential for life in Antarctic subglacial lakes. Hydrol. Process. 2003, 17, 795–814. [Google Scholar] [CrossRef]
- Siegert, M.J.; Carter, S.; Tabacco, I.; Popov, S.; Blankenship, D.D. A revised inventory of Antarctic subglacial lakes. Antarct. Sci. 2005, 17, 453–460. [Google Scholar] [CrossRef]
- Bell, R.E.; Studinger, M.; Shuman, C.A.; Fahnestock, M.A.; Joughin, I. Large subglacial lakes in East Antarctica at the onset of fast-flowing ice streams. Nature 2007, 445, 904–907. [Google Scholar] [CrossRef]
- Jones, N. Russians celebrate vostok victory. Nature 2012. [Google Scholar] [CrossRef]
- Siegert, M.J.; Clarke, R.J.; Mowlem, M.; Ross, N.; Hill, C.S.; Tait, A.; Hodgson, D.; Parnell, J.; Tranter, M.; Pearce, D.; et al. Clean access, measurement, and sampling of Ellsworth subglacial lake: A method for exploring deep Antarctic subglacial lake environments. Rev. Geophys. 2012, 50, RG1003. [Google Scholar] [CrossRef]
- Lake Ellsworth Consortium. Exploration of Ellsworth subglacial lake: A concept paper on the development, organisation and execution of an experiment to explore, measure and sample the environment of a West Antarctic subglacial lake. Rev. Env. Sci. Biotechnol. 2007, 6, 1569–1705.
- Pearce, D.A. Antarctic subglacial lake exploration: A new frontier in microbial ecology. ISME J. 2009, 3, 877–880. [Google Scholar] [CrossRef]
- Fricker, H.A.; Powell, R.; Priscu, J.; Tulaczyk, S.; Anandakrishnan, S.; Christner, B.; Fisher, A.T.; Holland, D.; Horgan, H.; Jacobel, R.; et al. Siple coast subglacial aquatic environments: The whillans ice stream subglacial access research drilling project. Geophys. Monogr. Ser. 2011, 194, 199–219. [Google Scholar]
- Hodgson, D.A.; Roberts, S.J.; Bentley, M.J.; Smith, J.A.; Johnson, J.S.; Verleyen, E.; Vyverman, W.; Hodson, A.J.; Leng, M.J.; Cziferszky, A.; et al. Exploring former subglacial Hodgson Lake, Antarctica paper i: Site description, geomorphology and limnology. Quat. Sci. Rev. 2009, 28, 2295–2309. [Google Scholar] [CrossRef]
- Schutte, U.M.; Abdo, Z.; Bent, S.J.; Williams, C.J.; Schneider, G.M.; Solheim, B.; Forney, L.J. Bacterial succession in a glacier foreland of the high Arctic. ISME J. 2009, 3, 1258–1268. [Google Scholar] [CrossRef]
- Anesio, A.M.; Hodson, A.J.; Fritz, A.; Psenner, R.; Sattler, B. High microbial activity on glaciers: Importance to the global carbon cycle. Glob. Chang. Biol. 2009, 15, 955–960. [Google Scholar] [CrossRef]
- Gaidos, E.; Lanoil, B.; Thorsteinsson, T.; Graham, A.; Skidmore, M.; Han, S.K.; Rust, T.; Popp, B. A viable microbial community in a subglacial volcanic crater lake, Iceland. Astrobiology 2004, 4, 327–344. [Google Scholar] [CrossRef]
- Miteva, V.I.; Brenchley, J.E. Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl. Environ. Microbiol. 2005, 71, 7806–7818. [Google Scholar] [CrossRef]
- Abyzov, S.S.; Hoover, R.B.; Mitskevich, I.N.; Mulyukin, A.L.; Poglazova, M.N.; Rozanov, A.Y. Microbiological Methodology in Astrobiology. Microbial extremophiles: psychrophiles i. In Astrobiology and Planetary Missions, Proceedings of the SPIE 2005, San Diego, CA, USA, 31 July 2005. [CrossRef]
- Gaidos, E.; Marteinsson, V.; Thorsteinsson, T.; Jóhannesson, T.; Rúnarsson, A.; Stefansson, A.; Glazer, B.; Lanoil, B.; Skidmore, M.; Han, S.; et al. An oligarchic microbial assemblage in the anoxic bottom waters of a volcanic subglacial lake. ISME J. 2009, 3, 486–497. [Google Scholar] [CrossRef]
- Marteinsson, V.T.; Rúnarsson, Á.; Stefánsson, A.; Thorsteinsson, T.; Jóhannesson, T.; Magnússon, S.H.; Reynisson, E.; Einarsson, B.; Wade, N.; Morrison, H.G.; et al. Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J. 2013, 7, 427–437. [Google Scholar] [CrossRef]
- Christner, B.C.; Royston-Bishop, G.; Foreman, C.M.; Arnold, B.R.; Tranter, M.; Welch, K.A.; Berry Lyons, W.; Tsapin, A.I.; Studinger, M.; Priscu, J.C. Limnological conditions in subglacial Lake Vostok, Antarctica. Limnol. Oceanogr. 2006, 51, 2485–2501. [Google Scholar] [CrossRef]
- D’Elia, T.; Veerapaneni, R.; Rogers, S.O. Isolation of microbes from Lake Vostok accretion ice. Appl. Environ. Microbiol. 2008, 74, 4962–4965. [Google Scholar] [CrossRef]
- D’Elia, T.; Veerapaneni, R.; Theraisnathan, V.; Rogers, S.O. Isolation of fungi from Lake Vostok accretion ice. Mycologia 2009, 101, 751–763. [Google Scholar] [CrossRef]
- Rogers, S.O.; Shtarkman, Y.M.S.; Koçer, Z.A.; Edgar, R.; Veerapaneni, R.; D’Elia, T. Ecology of subglacial Lake Vostok (Antarctica), based on metagenomic/metatranscriptomic analyses of accretion ice. Biology 2013, 2, 629–650. [Google Scholar] [CrossRef]
- Skidmore, M. Microbial communities in Antarctic subglacial aquatic environments (SAE). In Exploration and Study of Antarctic Subglacial Aquatic Environments; Siegert, M.J., Kennicutt II, M.C., Bindschandler, R.A., Eds.; ACU Press: Washington, DC, USA, 2011; AGU Geophysical Monograph Series; Volume 192, pp. 61–81. [Google Scholar]
- Bulat, S.A.; Alekhina, I.A.; Lipenkov, V.Y.; Lukin, V.V.; Marie, D.; Petit, J.R. Cell concentrations of microorganisms in glacial and lake ice of the vostok ice core, east antarctica. Microbiology 2009, 78, 808–810. [Google Scholar] [CrossRef]
- Priscu, J.C.; Adams, E.E.; Lyons, W.B.; Voytek, M.A.; Mogk, D.W.; Brown, R.L.; McKay, C.P.; Takacs, C.D.; Welch, K.A.; Wolf, C.F.; et al. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 1999, 286, 2141–2144. [Google Scholar] [CrossRef]
- Christner, B.C.; Mosley-Thompson, E.; Thompson, L.G.; Reeve, J.N. Isolation of bacteria and 16s RDNAs from Lake Vostok accretion ice. Environ. Microbiol. 2001, 3, 570–577. [Google Scholar] [CrossRef]
- Lanoil, B.; Skidmore, M.; Priscu, J.C.; Han, S.; Foo, W.; Vogel, S.W.; Tulaczyk, S.; Engelhardt, H. Bacteria beneath the West Antarctic ice sheet. Environ. Microbiol. 2009, 11, 609–615. [Google Scholar] [CrossRef]
- Hodgson, D.A.; Roberts, S.J.; Bentley, M.J.; Carmichael, E.L.; Smith, J.A.; Verleyen, E.; Vyverman, W.; Geissler, P.; Leng, M.J.; Sanderson, D.C.W. Exploring former subglacial Hodgson Lake, Antarctica. Paper ii: Palaeolimnology. Quat. Sci. Rev. 2009, 28, 2310–2325. [Google Scholar] [CrossRef]
- Porter, K.G.; Feig, Y.S. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 1980, 25, 943–948. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S RNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar]
- Stahl, D.A. Development and application of nucleic acid probes. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Ed.; John Wiley & Sons Ltd.: Chichester, UK, 1991; pp. 205–248. [Google Scholar]
- Manz, W.; Amann, R.; Ludwig, W.; Wagner, M.; Schleifer, K.H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria-problems and solutions. Syst. Appl. Microbiol. 1992, 15, 593–600. [Google Scholar] [CrossRef]
- Glockner, F.O.; Fuchs, B.M.; Amann, R. Bacterioplankton compositions of lakes and oceans: A first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 1999, 65, 3721–3726. [Google Scholar]
- Manz, W.; Amann, R.; Ludwig, W.; Vancanneyt, M.; Schleifer, K.H. Application of a suite of 16S RNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiol. UK 1996, 142, 1097–1106. [Google Scholar] [CrossRef]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jorgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623–626. [Google Scholar] [CrossRef]
- Glockner, F.O.; Zaichikov, E.; Belkova, N.; Denissova, L.; Pernthaler, J.; Pernthaler, A.; Amann, R. Comparative 16S RNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of Actinobacteria. Appl. Environ. Microbiol. 2000, 66, 5053–5065. [Google Scholar] [CrossRef]
- Meier, H.; Amann, R.; Ludwig, W.; Schleifer, K.H. Specific oligonucleotide probes for in situ detection of a major group of Gram-positive bacteria with low DNA G + C content. Syst. Appl. Microbiol. 1999, 22, 186–196. [Google Scholar] [CrossRef]
- Wallner, G.; Amann, R.; Beisker, W. Optimizing fluorescent in situ hybridization with RNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 1993, 14, 136–143. [Google Scholar] [CrossRef]
- Herrera, A.; Cockell, C.S.; Self, S.; Blaxter, M.; Reitner, J.; Arp, G.; Drose, W.; Thorsteinsson, T.; Tindle, A.G. Bacterial colonization and weathering of terrestrial obsidian in Iceland. Geomicrobiol. J. 2008, 25, 25–37. [Google Scholar] [CrossRef]
- Cockell, C.S.; Olsson, K.; Knowles, F.; Kelly, L.; Herrera, A.; Thorsteinsson, T.; Marteinsson, V. Bacteria in weathered basaltic glass, iceland. Geomicrobiol. J. 2009, 26, 491–507. [Google Scholar] [CrossRef]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; van Gijsegem, F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar]
- Ryan, F.J. Selected methods of neurospora genetics. Methods Med. Res. 1950, 3, 51–75. [Google Scholar]
- Geneious homepage. Available online: http://www.geneious.com (accessed on 1 July 2013).
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S RNA-based studies. PloS One 2011, 6, e27310. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Alvarez, V.; Teal, T.K.; Schmidt, T.M. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009, 3, 1314–1317. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics rast server-a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinforma. 2008, 9, 386:1–386:8. [Google Scholar]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glockner, F.O. Silva: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with arb. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S RNA gene database and workbench compatible with arb. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The ribosomal database project: Improved alignments and new tools for RNA analysis. Nucleic Acids Res. 2009, 37, D141–D145. [Google Scholar] [CrossRef]
- Chao, A. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Dong, H.; Zhang, G.; Jiang, H.; Yu, B.; Chapman, L.R.; Lucas, C.R.; Fields, M.W. Microbial diversity in sediments of saline Qinghai Lake, China: Linking geochemical controls to microbial ecology. Microb. Ecol. 2006, 51, 65–82. [Google Scholar] [CrossRef]
- Jiang, H.; Dong, H.; Zhang, G.; Yu, B.; Chapman, L.R.; Fields, M.W. Microbial diversity in water and sediment of Lake Chaka, an athalassohaline lake in Northwestern China. Appl. Environ. Microbiol. 2006, 72, 3832–3845. [Google Scholar] [CrossRef]
- Mancuso, C.A.; Franzmann, P.D.; Burton, H.R.; Nichols, P.D. Microbial community structure and biomass estimates of a methanogenic Antarctic lake ecosystem as determined by phospholipid analyses. Microb. Ecol. 1990, 19, 73–95. [Google Scholar] [CrossRef]
- Glockner, F.O.; Kube, M.; Bauer, M.; Teeling, H.; Lombardot, T.; Ludwig, W.; Gade, D.; Beck, A.; Borzym, K.; Heitmann, K.; et al. Complete genome sequence of the marine planctomycete Pirellula sp. Strain 1. Proc. Natl. Acad. Sci. USA 2003, 100, 8298–8303. [Google Scholar] [CrossRef]
- Breznak, J.A.; Canale-Parola, E. Spirochaeta aurantia, a pigmented, facultatively anaerobic spirochete. J. Bacteriol. 1969, 97, 386–395. [Google Scholar]
- Monciardini, P.; Cavaletti, L.; Schumann, P.; Rohde, M.; Donadio, S. Conexibacter woesei gen. Nov., sp. Nov., a novel representative of a deep evolutionary line of descent within the class actinobacteria. Int. J. Syst. Evol. Microbiol. 2003, 53, 569–576. [Google Scholar] [CrossRef]
- Aksenova, H.Y.; Rainey, F.A.; Janssen, P.H.; Zavarzin, G.A.; Morgan, H.W. Spirochaeta-thermophila sp-nov, an obligately anaerobic, polysaccharolytic, extremely thermophilic bacterium. Int. J. Syst. Bacteriol. 1992, 42, 175–177. [Google Scholar] [CrossRef]
- Botero, L.M.; Brown, K.B.; Brumefield, S.; Burr, M.; Castenholz, R.W.; Young, M.; McDermott, T.R. Thermobaculum terrenum gen. nov., sp. nov.: A non-phototrophic gram-positive thermophile representing an environmental clone group related to the chloroflexi (green non-sulfur bacteria) and thermomicrobia. Arch. Microbiol. 2004, 181, 269–277. [Google Scholar] [CrossRef]
- Tyson, G.W.; Lo, I.; Baker, B.J.; Allen, E.E.; Hugenholtz, P.; Banfield, J.F. Genome-directed isolation of the key nitrogen fixer Leptospirillum ferrodiazotrophum sp. nov. From an acidophilic microbial community. Appl. Environ. Microbiol. 2005, 71, 6319–6324. [Google Scholar] [CrossRef]
- Maymo-Gatell, X.; Anguish, T.; Zinder, S.H. Reductive dechlorination of chlorinated ethenes and 1, 2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 1999, 65, 3108–3113. [Google Scholar]
- Ward, N.; Larsen, O.; Sakwa, J.; Bruseth, L.; Khouri, H.; Durkin, A.S.; Dimitrov, G.; Jiang, L.; Scanlan, D.; Kang, K.H.; et al. Genomic insights into methanotrophy: The complete genome sequence of Methylococcus capsulatus (bath). PLoS Biol. 2004, 2, e303. [Google Scholar] [CrossRef] [Green Version]
- Shivaji, S.; Kumari, K.; Kishore, K.H.; Pindi, P.K.; Rao, P.S.; Radha Srinivas, T.N.; Asthana, R.; Ravindra, R. Vertical distribution of bacteria in a lake sediment from antarctica by culture-independent and culture-dependent approaches. Res. Microbiol. 2011, 162, 191–203. [Google Scholar] [CrossRef]
- Sjoling, S.; Cowan, D.A. High 16S RDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 2003, 7, 275–282. [Google Scholar] [CrossRef]
- Brambilla, E.; Hippe, H.; Hagelstein, A.; Tindall, B.J.; Stackebrandt, E. 16S RDNAdiversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, Mcmurdo Dry Valleys, Antarctica. Extremophiles 2001, 5, 23–33. [Google Scholar] [CrossRef]
- Sutherland, D.L.; Hawes, I. Annual growth layers as proxies of past growth conditions for benthic microbial mats in a perennially ice-covered Antarctic lake. FEMS Microbiol. Ecol. 2009, 67, 279–292. [Google Scholar] [CrossRef]
- Li, S.; Xiao, X.; Yin, X.; Wang, F. Bacterial community along a historic lake sediment core of Ardley Island, West Antarctica. Extremophiles 2006, 10, 461–467. [Google Scholar] [CrossRef]
- Bowman, J.P.; McCammon, S.A.; Gibson, J.A.; Robertson, L.; Nichols, P.D. Prokaryotic metabolic activity and community structure in Antarctic continental shelf sediments. Appl. Environ. Microbiol. 2003, 69, 2448–2462. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Brambilla, E.; Cousin, S.; Dirks, W.; Pukall, R. Culture-independent analysis of bacterial species from an anaerobic mat from Lake Fryxell, Antarctica: Prokaryotic diversity revisited. Cell. Mol. Biol. 2004, 50, 517–524. [Google Scholar]
- Purdy, K.J.; Nedwell, D.B.; Embley, T.M. Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 2003, 69, 3181–3191. [Google Scholar] [CrossRef]
- Bowman, J.P.; Rea, S.M.; McCammon, S.A.; McMeekin, T.A. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, eastern Antarctica. Environ. Microbiol. 2000, 2, 227–237. [Google Scholar] [CrossRef]
- Chen, M.; Xiao, X.; Wang, P.; Zeng, X.; Wang, F. Arthrobacter ardleyensis sp. nov., isolated from Antarctic Lake sediment and deep-sea sediment. Arch. Microbiol. 2005, 183, 301–305. [Google Scholar] [CrossRef]
- Wang, Z.W.; Liu, Y.H.; Dai, X.; Wang, B.J.; Jiang, C.Y.; Liu, S.J. Flavobacterium saliperosum sp. nov., isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 2006, 56, 439–442. [Google Scholar] [CrossRef]
- Karr, E.A.; Ng, J.M.; Belchik, S.M.; Sattley, W.M.; Madigan, M.T.; Achenbach, L.A. Biodiversity of methanogenic and other archaea in the permanently frozen Lake Fryxell, Antarctica. Appl. Environ. Microbiol. 2006, 72, 1663–1666. [Google Scholar] [CrossRef]
- Farmer, J.D.; Warner, N.H. Subglacial hydrothermal alteration minerals in jökulhlaup deposits of southern iceland, with implications for detecting past or present habitable environments on Mars. Astrobiology 2010, 10, 523–547. [Google Scholar] [CrossRef]
- Bjornsson, H. Subglacial lakes and jokulhlaups in Iceland. Glob. Planet. Change 2002, 35, 255–271. [Google Scholar] [CrossRef]
- Garchar, L.; Wendlandt, R.; Martini, B.; Owens, L. Geochemistry of a sub glacial volcanic hydrothermal system at Mount Spurr, Alaska. In Proceedings of Thirty Seventh Workshop on Geothermal Reservoir Engineering Stanford, Stanford, CA, USA, 30 January–1 February 2012.
- Petit, J.R.; Alekhina, I.A.; Bulat, S.A. A hydrothermal contribution to the Vostok subglacial lake (Antarctica) suggested from bacterial gene analysis and the stable isotope composition of deep ice core samples. In Proceedings of 35th COSPAR Scientific Assembly, Paris, France, 18–25 July 2004; p. 832.
- Lavire, C.; Normand, P.; Alekhina, I.; Bulat, S.; Prieur, D.; Birrien, J.-L.; Fournier, P.; Hänni, C.; Petit, J.-R. Presence of Hydrogenophilus thermoluteolus DNA in accretion ice in the subglacial Lake Vostok, Antarctica, assessed using rrs, cbb and hox. Environ. Microbiol. 2006, 8, 2106–2114. [Google Scholar] [CrossRef]
- Parkes, R.J.; Cragg, B.A.; Wellsbury, P. Recent studies on bacterial populations and processes in subseafloor sediments: A review. Hydrogeol. J. 2000, 8, 11–28. [Google Scholar] [CrossRef]
- Park, B.-J.; Park, S.-J.; Yoon, D.-N.; Schouten, S.; Sinninghe Damsté, J.S.; Rhee, S.-K. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl. Environ. Microbiol. 2010, 76, 7575–7587. [Google Scholar] [CrossRef]
- Mathis, B.J.; Marshall, C.W.; Milliken, C.E.; Makkar, R.S.; Creager, S.E.; May, H.D. Electricity generation by thermophilic microorganisms from marine sediment. Appl. Microbiol. Biotechnol. 2008, 78, 147–155. [Google Scholar] [CrossRef]
- Li, H.; Yu, Y.; Luo, W.; Zeng, Y.; Chen, B. Bacterial diversity in surface sediments from the Pacific Arctic Ocean. Extremophiles 2009, 13, 233–246. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Pearce, D.A.; Hodgson, D.A.; Thorne, M.A.S.; Burns, G.; Cockell, C.S. Preliminary Analysis of Life within a Former Subglacial Lake Sediment in Antarctica. Diversity 2013, 5, 680-702. https://doi.org/10.3390/d5030680
Pearce DA, Hodgson DA, Thorne MAS, Burns G, Cockell CS. Preliminary Analysis of Life within a Former Subglacial Lake Sediment in Antarctica. Diversity. 2013; 5(3):680-702. https://doi.org/10.3390/d5030680
Chicago/Turabian StylePearce, David A., Dominic A. Hodgson, Michael A. S. Thorne, Gavin Burns, and Charles S. Cockell. 2013. "Preliminary Analysis of Life within a Former Subglacial Lake Sediment in Antarctica" Diversity 5, no. 3: 680-702. https://doi.org/10.3390/d5030680
APA StylePearce, D. A., Hodgson, D. A., Thorne, M. A. S., Burns, G., & Cockell, C. S. (2013). Preliminary Analysis of Life within a Former Subglacial Lake Sediment in Antarctica. Diversity, 5(3), 680-702. https://doi.org/10.3390/d5030680




