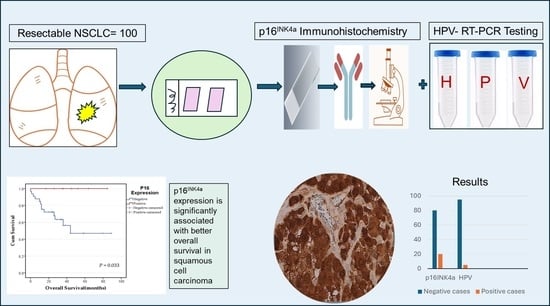
Human Papillomavirus Is Rare and Does Not Correlate with p16INK4A Expression in Non-Small-Cell Lung Cancer in a Jordanian Subpopulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. Extraction of DNA
2.3. HPV Detection and Genotyping
2.4. Immunohistochemistry (IHC)
2.5. p16INK4a Protein Expression Scoring
2.6. Statistical Analysis
3. Results
3.1. Patients’ Characteristics
3.2. HPV Detection and Correlation with p16INK4a Expression
3.3. p16INK4a Expression Detection and Correlation with the Clinicopathological Features
3.4. Survival Analysis of p16INK4a Expression and Other Clinicopathological Variables
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HPV | Human papillomavirus |
| HR-HPV | High-risk human papillomavirus |
| NSCLC | Non-small-cell lung cancer |
| IHC | Immunohistochemistry |
| FFPE | Formalin-fixed, paraffin-embedded |
| KHCC | King Hussein Cancer Center |
| RT-PCR | Real-time polymerase chain reaction |
| AJCC | American Joint Committee on Cancer protocol |
| ADC | Adenocarcinoma |
| SqCC | Squamous cell carcinoma |
| LR-HPV | Low-risk human papillomavirus |
| CDK | Cyclin-dependent kinase |
| WHO | World Health Organization |
| OS | Overall survival |
| DFS | Disease-free survival |
| EGFR | Epidermal growth factor receptor |
| ALK | Anaplastic lymphoma kinase |
| ISH | In situ hybridization |
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global Cancer Statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Sarchianaki, E.; Derdas, S.P.; Ntaoukakis, M.; Vakonaki, E.; Lagoudaki, E.D.; Lasithiotaki, I.; Sarchianaki, A.; Koutsopoulos, A.; Symvoulakis, E.K.; Spandidos, D.A.; et al. Detection and Genotype Analysis of Human Papillomavirus in Non-Small Cell Lung Cancer Patients. Tumor Biol. 2014, 35, 3203–3209. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Schiller, J.H.; Gazdar, A.F. Lung Cancer in Never Smokers—A Different Disease. Nat. Rev. Cancer 2007, 7, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Taioli, E.; Ragin, C.C. Human Papillomavirus Type 16 and 18 in Primary Lung Cancers—A Meta-Analysis. Carcinogenesis 2009, 30, 1722–1728. [Google Scholar] [CrossRef]
- Spyratos, D.; Zarogoulidis, P.; Porpodis, K.; Tsakiridis, K.; Machairiotis, N.; Katsikogiannis, N.; Kougioumtzi, I.; Dryllis, G.; Kallianos, A.; Rapti, A.; et al. Occupational Exposure and Lung Cancer. J. Thorac. Dis. 2013, 5 (Suppl. S4), S44–S45. [Google Scholar] [CrossRef]
- de Freitas, A.C.; Gurgel, A.P.; de Lima, E.G.; de França São Marcos, B.; do Amaral, C.M.M. Human Papillomavirus and Lung Cancinogenesis: An Overview. J. Cancer Res. Clin. Oncol. 2016, 142, 2415–2427. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Ding, J.; Shi, H.-Z. HPV and Lung Cancer Risk: A Meta-Analysis. J. Clin. Virol. 2015, 63, 84–90. [Google Scholar] [CrossRef]
- Bruni, L.; Albero, G.; Serrano, B.; Mena, M.; Collado, J.J.; Gómez, D.; Muñoz, J.; Bosch, F.X.; de Sanjosé, S. ICO/IARC Information Centre on HPV and Cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in the World. Summary Report 10 March 2023. Available online: https://hpvcentre.net/statistics/reports/XWX.pdf (accessed on 16 March 2024).
- Corneanu, L.M.; Stănculescu, D.; Corneanu, C. HPV and Cervical Squamous Intraepithelial Lesions: Clinicopathological Study. Rom. J. Morphol. Embryol. 2011, 52, 89–94. [Google Scholar] [PubMed]
- Berman, T.A.; Schiller, J.T. Human Papillomavirus in Cervical Cancer and Oropharyngeal Cancer: One Cause, Two Diseases. Cancer 2017, 123, 2219–2229. [Google Scholar] [CrossRef]
- Muñoz, N.; Castellsagué, X.; de González, A.B.; Gissmann, L. Chapter 1: HPV in the Etiology of Human Cancer. Vaccine 2006, 24, S1–S10. [Google Scholar] [CrossRef]
- Gillison, M.L.; Koch, W.M.; Capone, R.B.; Spafford, M.; Westra, W.H.; Wu, L.; Zahurak, M.L. Evidence for a Causal Association Between Human Papillomavirus and a Subset of Head and Neck Cancers. J. Natl. Cancer Inst. 2000, 92, 709–720. [Google Scholar] [CrossRef]
- Preti, M.; Boldorini, R.; Gallio, N.; Cavagnetto, C.; Borella, F.; Pisapia, E.; Ribaldone, R.; Bovio, E.; Bertero, L.; Airoldi, C.; et al. Human papillomavirus genotyping in high-grade vaginal intraepithelial neoplasia: A multicentric Italian study. J. Med. Virol. 2024, 96, e29474. [Google Scholar] [CrossRef]
- Sano, T.; Oyama, T.; Kashiwabara, K.; Fukuda, T.; Nakajima, T. Immunohistochemical Overexpression of P16 Protein Associated with Intact Retinoblastoma Protein Expression in Cervical Cancer and Cervical Intraepithelial Neoplasia. Pathol. Int. 1998, 48, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Doxtader, E.E.; Katzenstein, A.-L.A. The Relationship between P16 Expression and High-Risk Human Papillomavirus Infection in Squamous Cell Carcinomas from Sites Other than Uterine Cervix: A Study of 137 Cases. Hum. Pathol. 2012, 43, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Khleif, S.N.; DeGregori, J.; Yee, C.L.; Otterson, G.A.; Kaye, F.J.; Nevins, J.R.; Howley, P.M. Inhibition of Cyclin D-CDK4/CDK6 Activity Is Associated with an E2F-Mediated Induction of Cyclin Kinase Inhibitor Activity. Proc. Natl. Acad. Sci. USA 1996, 93, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; Jones, D.L. Human Papillomavirus Carcinogenesis: An Identity Crisis in the Retinoblastoma Tumor Suppressor Pathway. J. Virol. 2015, 89, 4708–4711. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin-Drubin, M.E.; Crum, C.P.; Münger, K. Human Papillomavirus E7 Oncoprotein Induces KDM6A and KDM6B Histone Demethylase Expression and Causes Epigenetic Reprogramming. Proc. Natl. Acad. Sci. USA 2011, 108, 2130–2135. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-W.; Wu, M.-F.; Wang, J.; Yeh, K.-T.; Goan, Y.-G.; Chiou, H.-L.; Chen, C.-Y.; Lee, H. Human Papillomavirus 16/18 E6 Oncoprotein Is Expressed in Lung Cancer and Related with P53 Inactivation. Cancer Res. 2007, 67, 10686–10693. [Google Scholar] [CrossRef]
- Syrjänen, K.; Syrjänen, S.; Kellokoski, J.; Kärjä, J.; Mäntyjärvi, R. Human Papillomavirus (HPV) Type 6 and 16 DNA Sequences in Bronchial Squamous Cell Carcinomas Demonstrated by in Situ DNA Hybridization. Lung 1989, 167, 33–42. [Google Scholar] [CrossRef]
- Bishop, J.A.; Ogawa, T.; Chang, X.; Illei, P.B.; Gabrielson, E.; Pai, S.I.; Westra, W.H. HPV Analysis in Distinguishing Second Primary Tumors From Lung Metastases in Patients With Head and Neck Squamous Cell Carcinoma. Am. J. Surg. Pathol. 2012, 36, 142–148. [Google Scholar] [CrossRef]
- Myong, N.-H. Cyclin D1 Overexpression, P16 Loss, and pRb Inactivation Play a Key Role in Pulmonary Carcinogenesis and Have a Prognostic Implication for the Long-Term Survival in Non-Small Cell Lung Carcinoma Patients. Cancer Res. Treat. 2008, 40, 45–52. [Google Scholar] [CrossRef]
- Sterlacci, W.; Tzankov, A.; Veits, L.; Zelger, B.; Bihl, M.P.; Foerster, A.; Augustin, F.; Fiegl, M.; Savic, S. A Comprehensive Analysis of P16 Expression, Gene Status, and Promoter Hypermethylation In Surgically Resected Non-Small Cell Lung Carcinomas. J. Thorac. Oncol. 2011, 6, 1649–1657. [Google Scholar] [CrossRef]
- An, H.J.; Koh, H.M.; Song, D.H. New P16 Expression Criteria Predict Lymph Node Metastasis in Patients With Non-Small Cell Lung Cancer. Vivo 2019, 33, 1885–1892. [Google Scholar] [CrossRef] [PubMed]
- Pezzuto, A.; Cappuzzo, F.; D’Arcangelo, M.; Ciccozzi, M.; Navarini, L.; Guerrini, S.; Ricci, A.; D’Ascanio, M.; Carico, E. Prognostic Value of P16 Protein in Patients With Surgically Treated Non-Small Cell Lung Cancer; Relationship With Ki-67 and PD-L1. Anticancer Res. 2020, 40, 983–990. [Google Scholar] [CrossRef] [PubMed]
- Qaqish, A.; Abdo, N.; Abbas, M.M.; Saadeh, N.; Alkhateeb, M.; Msameh, R.; Tarawneh, S.; Al-Masri, M. Awareness and Knowledge of Physicians and Residents on the Non-Sexual Routes of Human Papilloma Virus (HPV) Infection and Their Perspectives on Anti-HPV Vaccination in Jordan. PLoS ONE 2023, 18, e0291643. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Mirsadraee, S.; Oswal, D.; Alizadeh, Y.; Caulo, A.; van Beek, E.J. The 7th Lung Cancer TNM Classification and Staging System: Review of the Changes and Implications. World J. Radiol. 2012, 4, 128–134. [Google Scholar] [CrossRef]
- Detterbeck, F.C. The Eighth Edition TNM Stage Classification for Lung Cancer: What Does It Mean on Main Street? J. Thorac. Cardiovasc. Surg. 2018, 155, 356–359. [Google Scholar] [CrossRef]
- Würdemann, N.; Wagner, S.; Sharma, S.J.; Prigge, E.-S.; Reuschenbach, M.; Gattenlöhner, S.; Klussmann, J.P.; Wittekindt, C. Prognostic Impact of AJCC/UICC 8th Edition New Staging Rules in Oropharyngeal Squamous Cell Carcinoma. Front. Oncol. 2017, 7, 129. [Google Scholar] [CrossRef]
- Machczyński, P.; Majchrzak, E.; Niewinski, P.; Marchlewska, J.; Golusiński, W. A Review of the 8th Edition of the AJCC Staging System for Oropharyngeal Cancer According to HPV Status. Eur. Arch. Otorhinolaryngol. 2020, 277, 2407–2412. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-I.; Taki, T.; Higashiyama, M.; Kohno, N.; Miyake, M. P16 Protein Expression Is Associated with a Poor Prognosis in Squamous Cell Carcinoma of the Lung. Br. J. Cancer 2000, 82, 374–380. [Google Scholar] [CrossRef]
- Syrjänen, K. Detection of Human Papillomavirus in Lung Cancer: Systematic Review and Meta-Analysis. Anticancer Res. 2012, 32, 3235–3250. [Google Scholar] [PubMed]
- Nadji, S.A.; Mokhtari-Azad, T.; Mahmoodi, M.; Yahyapour, Y.; Naghshvar, F.; Torabizadeh, J.; Ziaee, A.A.; Nategh, R. Relationship between Lung Cancer and Human Papillomavirus in North of Iran, Mazandaran Province. Cancer Lett. 2007, 248, 41–46. [Google Scholar] [CrossRef]
- Hussen, B.M.; Ahmadi, G.; Marzban, H.; Fard Azar, M.E.; Sorayyayi, S.; Karampour, R.; Nahand, J.S.; Hidayat, H.J.; Moghoofei, M. The Role of HPV Gene Expression and Selected Cellular MiRNAs in Lung Cancer Development. Microb. Pathog. 2021, 150, 104692. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Amin Kotb, W.F.M.; Petersen, I. Incidence of Human Papilloma Virus in Lung Cancer. Lung Cancer 2009, 65, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Ando, M.; Kubo, A.; Isa, S.; Yamamoto, S.; Tsujino, K.; Kurata, T.; Ou, S.-H.I.; Takada, M.; Kawaguchi, T. Human Papilloma Virus in Non-Small Cell Lung Cancer in Never Smokers: A Systematic Review of the Literature. Lung Cancer 2014, 83, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, N.; Wang, A.; Kohler, D.; Santos, G.D.C.; Sykes, J.; Xu, J.; Pintilie, M.; Tsao, M.-S. Human Papilloma Virus Genome Is Rare in North American Non-Small Cell Lung Carcinoma Patients. Lung Cancer 2013, 79, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Joh, J.; Jenson, A.B.; Moore, G.D.; Rezazedeh, A.; Slone, S.P.; Ghim, S.; Kloecker, G.H. Human Papillomavirus (HPV) and Merkel Cell Polyomavirus (MCPyV) in Non Small Cell Lung Cancer. Exp. Mol. Pathol. 2010, 89, 222–226. [Google Scholar] [CrossRef]
- Goto, A.; Li, C.-P.; Ota, S.; Niki, T.; Ohtsuki, Y.; Kitajima, S.; Yonezawa, S.; Koriyama, C.; Akiba, S.; Uchima, H.; et al. Human Papillomavirus Infection in Lung and Esophageal Cancers: Analysis of 485 Asian Cases. J. Med. Virol. 2011, 83, 1383–1390. [Google Scholar] [CrossRef]
- Silva, E.M.; Mariano, V.S.; Pastrez, P.R.A.; Pinto, M.C.; Nunes, E.M.; Sichero, L.; Villa, L.L.; Scapulatempo-Neto, C.; Syrjanen, K.J.; Longatto-Filho, A. Human Papillomavirus Is Not Associated to Non-Small Cell Lung Cancer: Data from a Prospective Cross-Sectional Study. Infect. Agent. Cancer 2019, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.Y.; Keeney, M.; Law, M.; Donovan, J.; Aubry, M.-C.; Garcia, J. Detection of Human Papillomavirus in Non–Small Cell Carcinoma of the Lung. Hum. Pathol. 2015, 46, 1592–1597. [Google Scholar] [CrossRef] [PubMed]
- Iwakawa, R.; Kohno, T.; Enari, M.; Kiyono, T.; Yokota, J. Prevalence of Human Papillomavirus 16/18/33 Infection and P53 Mutation in Lung Adenocarcinoma. Cancer Sci. 2010, 101, 1891–1896. [Google Scholar] [CrossRef] [PubMed]
- Khasawneh, A.I.; Asali, F.F.; Kilani, R.M.; Abu-Raideh, J.A.; Himsawi, N.M.; Salameh, M.A.; Al Ghabbiesh, G.H.; Saleh, T. Prevalence and Genotype Distribution of Human Papillomavirus Among a Subpopulation of Jordanian Women. Int. J. Womens Health 2020, 9, 017–023. [Google Scholar] [CrossRef]
- Ragin, C.; Obikoya-Malomo, M.; Kim, S.; Chen, Z.; Flores-Obando, R.; Gibbs, D.; Koriyama, C.; Aguayo, F.; Koshiol, J.; Caporaso, N.E.; et al. HPV-Associated Lung Cancers: An International Pooled Analysis. Carcinogenesis 2014, 35, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Höti, N.; Ao, M.; Zhang, Z.; Zhu, H.; Li, L.; Askin, F.; Gabrielson, E.; Zhang, H.; Li, Q.K. Expression of P16 and P53 in Non-Small-Cell Lung Cancer: Clinicopathological Correlation and Potential Prognostic Impact. Biomark. Med. 2019, 13, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.G.; Zacharatos, P.; Kotsinas, A.; Liloglou, T.; Kyroudi, A.; Veslemes, M.; Rassidakis, A.; Halazonetis, T.D.; Field, J.K.; Kittas, C. Alterations of the P16-pRb Pathway and the Chromosome Locus 9p21–22 in Non-Small-Cell Lung Carcinomas. Am. J. Pathol. 1998, 153, 1749–1765. [Google Scholar] [CrossRef]
- Romagosa, C.; Simonetti, S.; López-Vicente, L.; Mazo, A.; Lleonart, M.E.; Castellvi, J.; Ramon Y Cajal, S. p16Ink4a Overexpression in Cancer: A Tumor Suppressor Gene Associated with Senescence and High-Grade Tumors. Oncogene 2011, 30, 2087–2097. [Google Scholar] [CrossRef]
- Paez-Ribes, M.; González-Gualda, E.; Doherty, G.J.; Muñoz-Espín, D. Targeting Senescent Cells in Translational Medicine. EMBO Mol. Med. 2019, 11, e10234. [Google Scholar] [CrossRef]
- Domen, A.; Deben, C.; De Pauw, I.; Hermans, C.; Lambrechts, H.; Verswyvel, J.; Siozopoulou, V.; Pauwels, P.; Demaria, M.; Van De Wiel, M.; et al. Prognostic Implications of Cellular Senescence in Resected Non-Small Cell Lung Cancer. Transl. Lung Cancer Res. 2022, 11, 1526–1539. [Google Scholar] [CrossRef]
- Saleh, T.; Bloukh, S.; Hasan, M.; Al Shboul, S. Therapy-Induced Senescence as a Component of Tumor Biology: Evidence from Clinical Cancer. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188994. [Google Scholar] [CrossRef] [PubMed]
- Robinson, L.A.; Jaing, C.J.; Pierce Campbell, C.; Magliocco, A.; Xiong, Y.; Magliocco, G.; Thissen, J.B.; Antonia, S. Molecular Evidence of Viral DNA in Non-Small Cell Lung Cancer and Non-Neoplastic Lung. Br. J. Cancer 2016, 115, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Marcos, B.F.S.; De Oliveira, T.H.A.; Do Amaral, C.M.M.; Muniz, M.T.C.; Freitas, A.C. Correlation between HPV PCNA, P16, and P21 Expression in Lung Cancer Patients. Cell. Microbiol. 2022, 2022, 9144334. [Google Scholar] [CrossRef]
- Bian, C.; Li, Z.; Xu, Y.; Wang, J.; Xu, L.; Shen, H. Clinical Outcome and Expression of Mutant P53, P16, and Smad4 in Lung Adenocarcinoma: A Prospective Study. World J. Surg. Oncol. 2015, 13, 128. [Google Scholar] [CrossRef]



| Variables | Number (%) | p16INK4a Positive b | p16INK4a Negative | p Value * |
|---|---|---|---|---|
| Total | 100 (100.0) | 20 (20.0) | 80 (80.0) | |
| Age (Years) | 0.232 | |||
| ≤60 | 23 (23.0) | 7 (35.0) | 16 (20.0) | |
| >60 | 77 (77.0) | 13 (65.0) | 64 (80.0) | |
| Gender | 0.072 | |||
| Male | 85 (85.0) | 14 (70.0) | 71 (88.7) | |
| Female | 15 (15.0) | 6 (30.0) | 9 (11.3) | |
| Smoking history a | 0.729 | |||
| Current/former smoker | 84 (84.8) | 16 (80.0) | 68 (86.1) | |
| Never smoker | 15 (15.2) | 4 (20.0) | 11 (13.9) | |
| Histological subtype | 1.000 | |||
| Adenocarcinoma (ADC) | 59 (59.0) | 12 (60.0) | 47 (58.8) | |
| Squamous cell carcinoma (SqCC) | 41 (41.0) | 8 (40.0) | 33 (41.2) | |
| Grade | 0.617 | |||
| Low-grade (well and moderately differentiated) | 41 (41.0) | 7 (35.0) | 34 (42.5) | |
| High-grade (poorly differentiated) | 59 (59.0) | 13 (65.0) | 46 (57.5) | |
| Tumor size | 0.193 | |||
| ≤3 cm | 36 (36.0) | 10 (50.0) | 26 (32.5) | |
| >3 cm | 64 (64.0) | 10 (50.0) | 54 (67.5) | |
| Lymph nodes metastasis | 0.799 | |||
| Positive | 37 (37.0) | 8 (40.0) | 29 (36.3) | |
| Negative | 63 (63.0) | 12 (60.0) | 51 (63.7) | |
| Pathological stage | 0.029 | |||
| I, II | 69 (69.0) | 18 (90.0) | 51 (63.7) | |
| III, IV | 31 (31.0) | 2 (10.0) | 29 (36.3) | |
| Predominant histological pattern (For ADC cases, 59 cases) | 0.422 | |||
| Lepidic | 14 (23.7) | 3 (25.0) | 11 (23.4) | |
| Acinar | 28 (47.5) | 4 (33.3) | 24 (51.1) | |
| Micropapillary | 2 (3.4) | 1 (8.3) | 1 (2.1) | |
| Solid | 15 (25.4) | 4 (33.3) | 11 (23.4) | |
| Papillary | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Recurrence/Progression | 1.000 | |||
| Present | 34 (34.0) | 7 (35.0) | 27 (33.8) | |
| Absent | 66 (66.0) | 13 (65.0) | 53 (66.3) | |
| HPV Status | ||||
| Positive | 5 (5.0) | 0 (0.0) | 5 (6.3) | 0.580 |
| Negative | 95 (95.0) | 20 (100.0) | 75 (93.7) |
| HPV-Positive Tumor | Age | Gender | Smoking History | Histological Subtype | Grade a | Pathological Stage | Recurrence/Progression | p16INK4a Expression b |
|---|---|---|---|---|---|---|---|---|
| HPV 16+ | 56 | Female | Former smoker | ADC | High | Stage I | Absent | Negative |
| HPV 18+ | 74 | Male | Smoker | ADC | Low | Stage II | Present | Negative |
| HPV 26+ | 72 | Male | Smoker | SqCC | Low | Stage I | Absent | Negative |
| HPV 26+ | 78 | Male | Smoker | ADC | Low | Stage III | Absent | Negative |
| HPV 52+ | 70 | Female | Non-smoker | ADC | Low | Stage I | Absent | Negative |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu Al Karsaneh, O.; Al Anber, A.; AlMustafa, S.; AlMa’aitah, H.; AlQadri, B.; Igbaria, A.; Tayem, R.; Khasawneh, M.; Batayha, S.; Saleh, T.; et al. Human Papillomavirus Is Rare and Does Not Correlate with p16INK4A Expression in Non-Small-Cell Lung Cancer in a Jordanian Subpopulation. Medicina 2024, 60, 660. https://doi.org/10.3390/medicina60040660
Abu Al Karsaneh O, Al Anber A, AlMustafa S, AlMa’aitah H, AlQadri B, Igbaria A, Tayem R, Khasawneh M, Batayha S, Saleh T, et al. Human Papillomavirus Is Rare and Does Not Correlate with p16INK4A Expression in Non-Small-Cell Lung Cancer in a Jordanian Subpopulation. Medicina. 2024; 60(4):660. https://doi.org/10.3390/medicina60040660
Chicago/Turabian StyleAbu Al Karsaneh, Ola, Arwa Al Anber, Sahar AlMustafa, Hussien AlMa’aitah, Batool AlQadri, Abir Igbaria, Rama Tayem, Mustafa Khasawneh, Shaima Batayha, Tareq Saleh, and et al. 2024. "Human Papillomavirus Is Rare and Does Not Correlate with p16INK4A Expression in Non-Small-Cell Lung Cancer in a Jordanian Subpopulation" Medicina 60, no. 4: 660. https://doi.org/10.3390/medicina60040660