Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea
Abstract
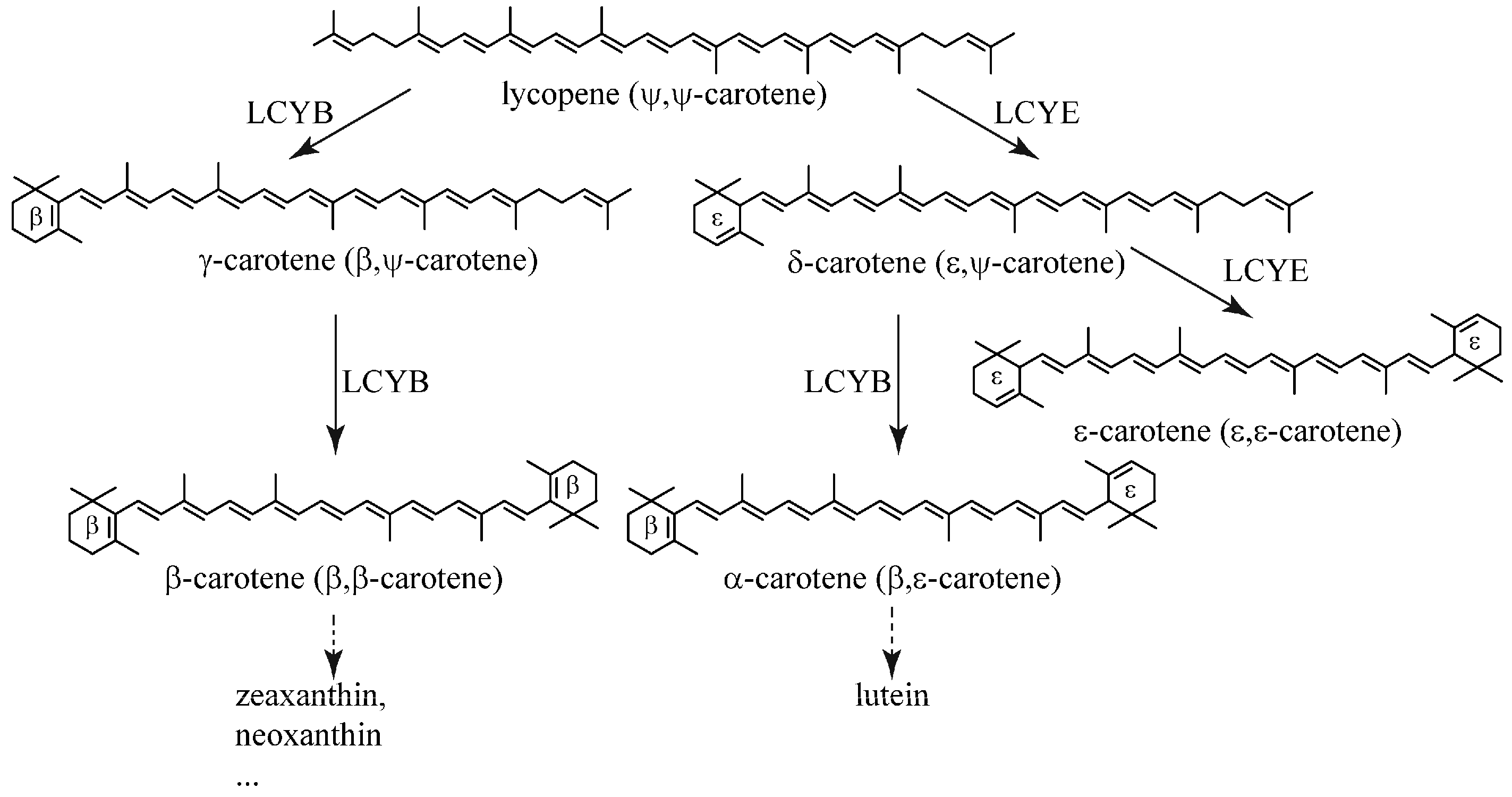
:1. Introduction
2. Results
2.1. BfLCYB Is an Intron-Less LCY Gene
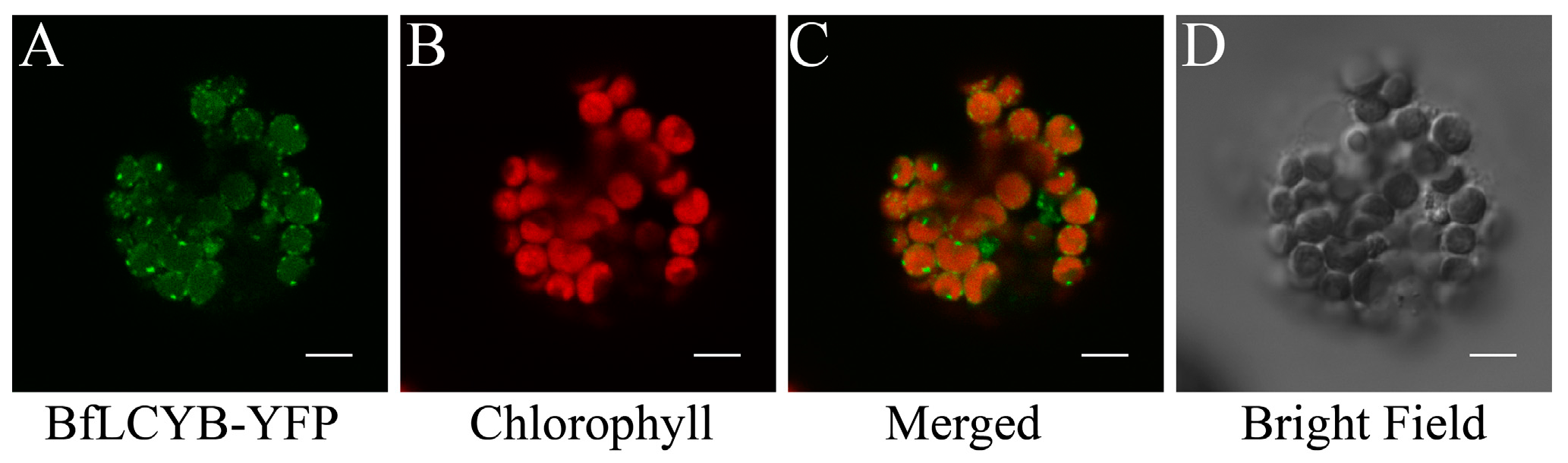
2.2. BfLCYB Localizes in Chloroplasts
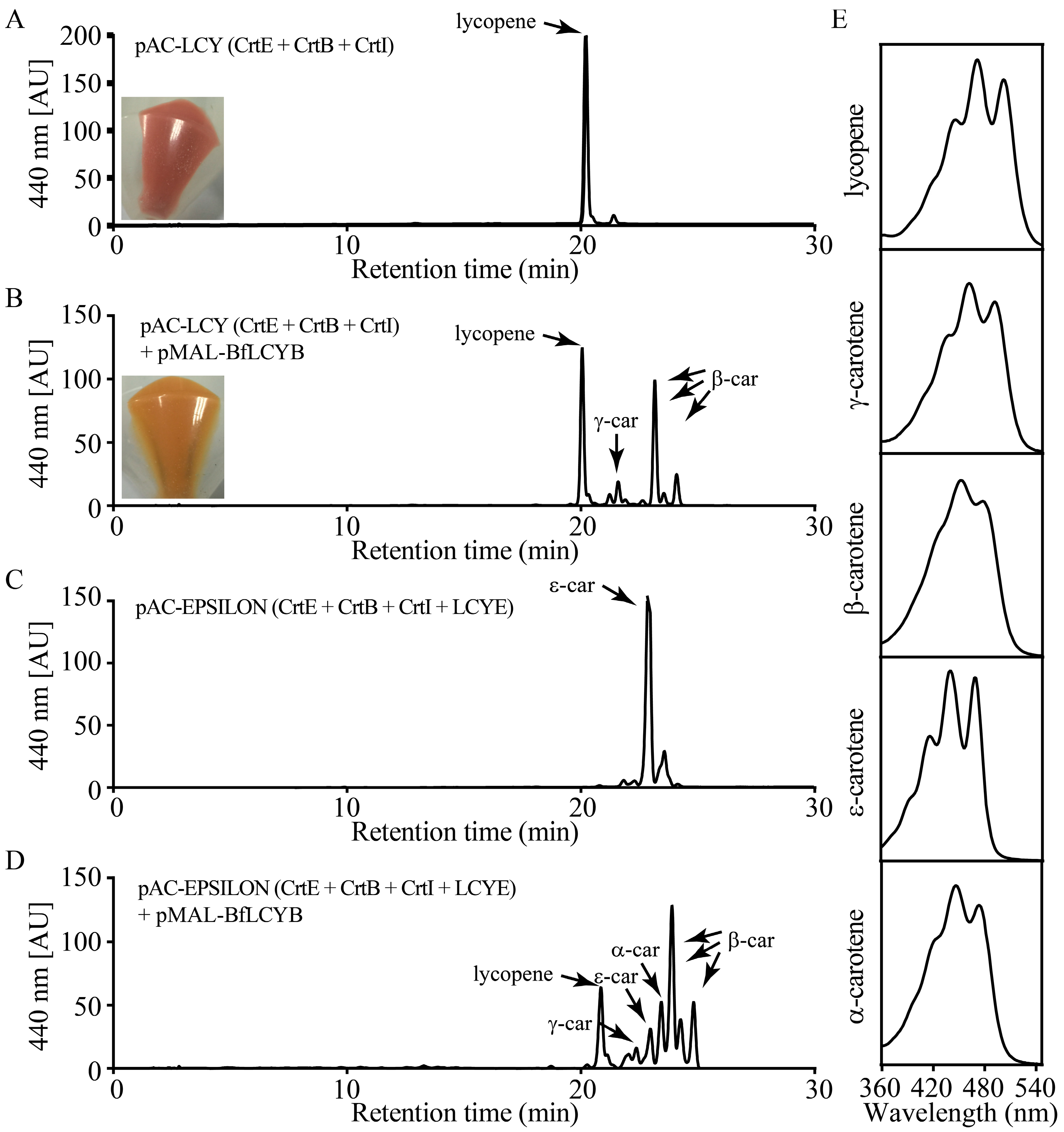
2.3. BfLCYB Is a Bicyclic Lycopene β-Cyclase
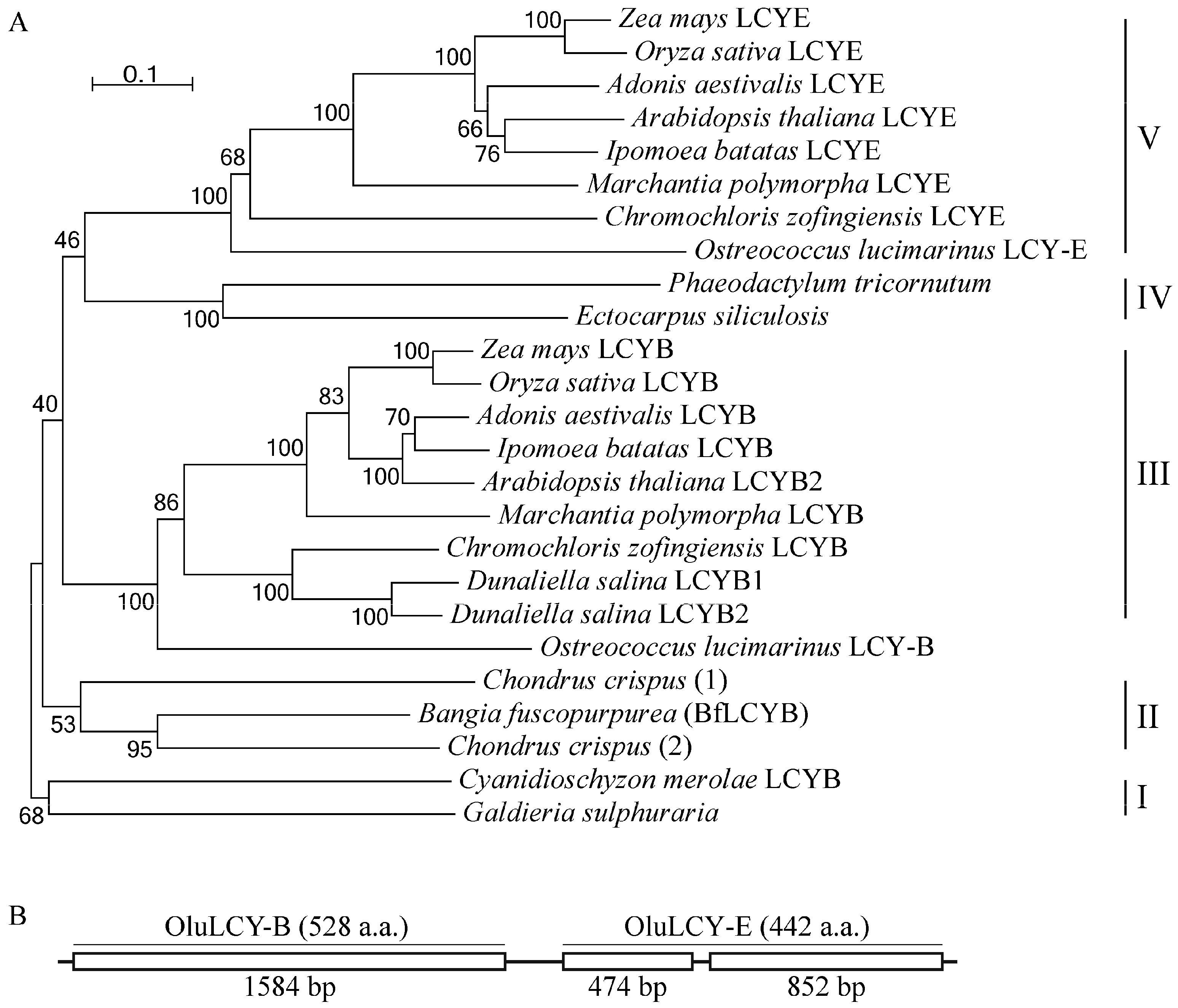
2.4. BfLCYB Is an Ancient Type of Lycopene Cyclase in Plants
3. Discussion
4. Materials and Methods
4.1. Material and Growth Condition
4.2. Sequence Analysis
4.3. Subcellular Localization
4.4. Functional Characterization
4.5. Carotenoid Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef] [PubMed]
- Takaichi, S. Carotenoids in algae: Distributions, biosyntheses and functions. Mar. Drugs 2011, 9, 1101–1118. [Google Scholar] [CrossRef] [PubMed]
- Britton, G.; Liaaen-Jensen, S.; Pfander, H. Carotenoids: Handbook; Birkhäuser: Basel, Switzerland, 2004. [Google Scholar]
- Cazzonelli, C.I. Carotenoids in nature: Insights from plants and beyond. Funct. Plant Biol. 2011, 38, 833–847. [Google Scholar] [CrossRef]
- Al-Babili, S.; Bouwmeester, H.J. Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 2015, 66, 161–186. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.L.; Li, L.; Shah, S.N.M.; Gong, Z.H. The relationship between red fruit colour formation and key genes of capsanthin biosynthesis pathway in Capsicum annuum. Biol. Plant. 2015, 59, 507–513. [Google Scholar] [CrossRef]
- Clotault, J.; Peltier, D.; Berruyer, R.; Thomas, M.; Briard, M.; Geoffriau, E. Expression of carotenoid biosynthesis genes during carrot root development. J. Exp. Bot. 2008, 59, 3563–3573. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X., Jr.; Gantt, E. Elucidation of the pathway to astaxanthin in the flowers of Adonis aestivalis. Plant Cell 2011, 23, 3055–3069. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Sola, M.Á.; Rodríguez-Concepción, M. Carotenoid biosynthesis in Arabidopsis: A colorful pathway. Arabidopsis Book 2012, 10, e0158. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G. Molecular evolution of carotenoid biosynthesis from bacteria to plants. Physiol. Plant. 2002, 116, 431–440. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Gantt, E. One ring or two? Determination of ring number in carotenoids by lycopene ε-cyclases. Proc. Natl. Acad. Sci. USA 2001, 98, 2905–2910. [Google Scholar] [CrossRef] [PubMed]
- Harjes, C.E.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T.; et al. Natural genetic variation in Lycopene Epsilon Cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Moise, A.R.; Al-Babili, S.; Wurtzel, E.T. Mechanistic aspects of carotenoid biosynthesis. Chem. Rev. 2014, 114, 164–193. [Google Scholar] [CrossRef] [PubMed]
- Römer, S. Carotenoids in higher plants and algae. In The Chloroplast: From Molecular Biology to Biotechnology; Argyroudi-Akoyunoglou, J.H., Senger, H., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 217–223. [Google Scholar]
- Schubert, N.; García-Mendoza, E.; Pacheco-Ruiz, I. Carotenoid composition of marine red algae. J. Phycol. 2006, 42, 1208–1216. [Google Scholar] [CrossRef]
- Takaichi, S.; Yokoyama, A.; Uchida, H.; Murakami, A. Carotenogenesis diversification in phylogenetic lineages of Rhodophyta. J. Phycol. 2016, 52, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X., Jr.; Lee, H.; Gantt, E. Carotenoid biosynthesis in the primitive red alga Cyanidioschyzon merolae. Eukaryot. Cell 2007, 6, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, N.J. Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology 2000, 26, 386–404. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Briesemeister, S.; Rahnenfuhrer, J.; Kohlbacher, O. YLoc—An interpretable web server for predicting subcellular localization. Nucleic Acids Res. 2010, 38, W497–W502. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Von Heijne, G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999, 8, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.X., Jr.; Gantt, E. A portfolio of plasmids for identification and analysis of carotenoid pathway enzymes: Adonis aestivalis as a case study. Photosynth. Res. 2007, 92, 245–259. [Google Scholar] [CrossRef] [PubMed]
- Blatt, A.; Bauch, M.E.; Porschke, Y.; Lohr, M. A lycopene β-cyclase/lycopene ε-cyclase/light-harvesting complex-fusion protein from the green alga Ostreococcus lucimarinus can be modified to produce α-carotene and β-carotene at different ratios. Plant J. 2015, 82, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Lohr, M.; Im, C.S.; Grossman, A.R. Genome-based examination of chlorophyll and carotenoid biosynthesis in Chlamydomonas reinhardtii. Plant Physiol. 2005, 138, 490–515. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, M. Carotenoid biosynthesis in diatoms. Photosynth. Res. 2010, 106, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Coesel, S.; Oborník, M.; Varela, J.; Falciatore, A.; Bowler, C. Evolutionary origins and functions of the carotenoid biosynthetic pathway in marine diatoms. PLoS ONE 2008, 3, e2896. [Google Scholar] [CrossRef] [PubMed]
- Kuczynska, P.; Jemiola-Rzeminska, M.; Strzalka, K. Photosynthetic pigments in diatoms. Mar. Drugs 2015, 13, 5847–5881. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-J.; Zhu, J.-Y.; Xu, P.; Xu, J.-R.; Lin, X.-Z.; Huang, C.-K.; Song, W.-L.; Peng, G.; Wang, G.-C. Characterization of the life history of Bangia fuscopurpurea (Bangiaceae, Rhodophyta) in connection with its cultivation in China. Aquaculture 2008, 278, 101–109. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-E.; Jin, Q.-P.; Xiao, Y.; Xu, P.; Lu, S. Improved methods for basic molecular manipulation of the red alga Porphyra umbilicalis (Rhodophyta: Bangiales). J. Appl. Phycol. 2012, 25, 245–252. [Google Scholar] [CrossRef]
- Ramos, A.; Coesel, S.; Marques, A.; Rodrigues, M.; Baumgartner, A.; Noronha, J.; Rauter, A.; Brenig, B.; Varela, J. Isolation and characterization of a stress-inducible Dunaliella salina Lcy-β gene encoding a functional lycopene β-cyclase. Appl. Microbiol. Biotechnol. 2008, 79, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-E.; Huang, X.-Q.; Hang, Y.; Deng, Y.-Y.; Lu, Q.-Q.; Lu, S. The P450-type carotene hydroxylase PuCHY1 from Porphyra suggests the evolution of carotenoid metabolism in red algae. J. Integr. Plant Biol. 2014, 56, 902–915. [Google Scholar] [CrossRef] [PubMed]
- Norris, S.R.; Barrette, T.R.; DellaPenna, D. Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 1995, 7, 2139–2149. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′-3′) 1,2 |
|---|---|
| DF1 | MMNAAYTAYGGNKKNTGGBWNGAYGAR |
| DR1 | GGRTGNACSADNSHNGCNGYNSCNCCNAWNSC |
| DF2 | ATGBTNYTNATGGAYKDNMGNGA |
| DR2 | RNVNNCCNCCNAYNGGDATNWV |
| RR1 | GGCCCCCCCATGGGGATCAGACAAAACTC |
| RR2 | CAGACAAAACTCCTCATCGAGCACCCGC |
| RR3 | CACCCGCGTCACGGTGATGCCGTC |
| RF1 | GGTGCTGATGGACTACCGGGATGG |
| RF2 | GGGATGGGCACATGCAGGGGGAC |
| RF3 | GGACGCCGCGGGGCGGGCCGAGTC |
| HF | ATGGAGGCGTTCATCCCCGCCTC |
| ER | CTACGCCTCGTCCTCTGGCAGCGG |
| gHF | AATCAGTCAGTTTCGGTGATCTTTC |
| gER | CCGCCGAAACGGCCTACCCCT |
| SpeI-LCY-F | GACTAGTATGGAGGCGTTCATCCCCGCCTC |
| SpeI-LCY-R | GGGACTAGTCTACGCCTCGTCCTCTGGCAGCGG |
| BamHI-LCY-F | GGATCCATGGAGGCGTTCATCCCCGCCTC |
| BamHI-LCY-R | GGGGATCCCTACGCCTCGTCCTCTGGCAGCGG |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, T.-J.; Huang, X.-Q.; Qu, Y.-Y.; Zhuang, Z.; Deng, Y.-Y.; Lu, S. Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea. Mar. Drugs 2017, 15, 116. https://doi.org/10.3390/md15040116
Cao T-J, Huang X-Q, Qu Y-Y, Zhuang Z, Deng Y-Y, Lu S. Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea. Marine Drugs. 2017; 15(4):116. https://doi.org/10.3390/md15040116
Chicago/Turabian StyleCao, Tian-Jun, Xing-Qi Huang, Yuan-Yuan Qu, Zhong Zhuang, Yin-Yin Deng, and Shan Lu. 2017. "Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea" Marine Drugs 15, no. 4: 116. https://doi.org/10.3390/md15040116
APA StyleCao, T.-J., Huang, X.-Q., Qu, Y.-Y., Zhuang, Z., Deng, Y.-Y., & Lu, S. (2017). Cloning and Functional Characterization of a Lycopene β-Cyclase from Macrophytic Red Alga Bangia fuscopurpurea. Marine Drugs, 15(4), 116. https://doi.org/10.3390/md15040116







