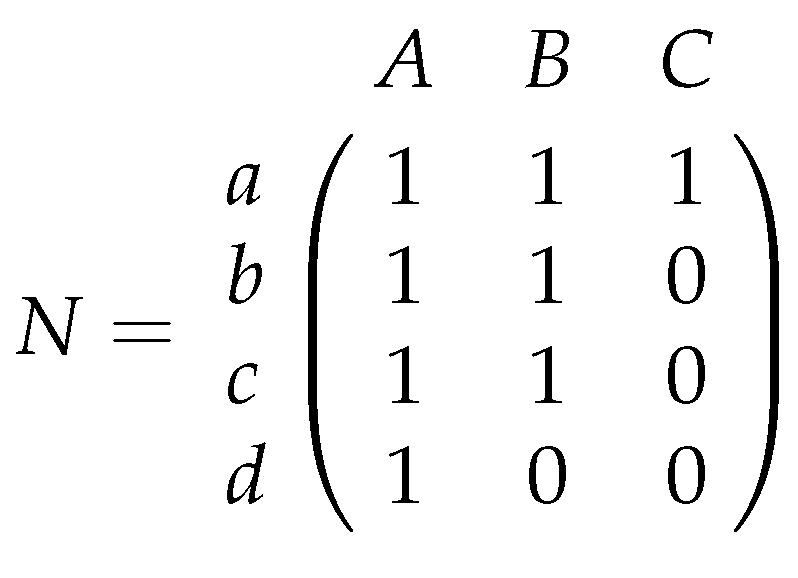1. Introduction
In applied fields like ecology, a
-matrix
M often represents the presence or absence of certain relationships between species or objects, for example, which bee species pollinates which plants or the occurrence and absence of species on several islands.
M is often called a
contingency table. Such a matrix
M can represent a bipartite graph (A bipartite graph
consists of two disjoint vertex sets
U and
V such that all edges are of the form
, where
and
). Connections of bees and plants are given by edges. Matrix
M is then called a
bi-adjacency matrix. (For a bipartite graph
with
and
, we construct its
bi-adjacency matrix M as follows: rows
represent vertices
in
U, and columns
correspond to vertices
in
If edge
, then we set
; otherwise, we have
) Ecologists call a matrix
nested if we find for every row pair
in
M the following condition; if
i has equal or a smaller number of 1 s than
j then
if and only if
Consider in
Figure 1 the nested matrix
N that represents the relationships between bee and plant species, and their corresponding bipartite graph.
The term
nestedness has already been introduced in 1937 by Hultén [
1]. A comprehensive history, discussion, and a definition of nestedness is given by Ulrich et al. [
2]. They describe nestedness as follows: “In a nested pattern, the species composition of small assemblages is a nested subset of the species composition of large assemblages”. Interpreting nestedness regarding biological properties in networks is quite interesting. In matrix
N of
Figure 1 bees b, c, and d pollinate less plants than bee a. However, it does not happen that these bees pollinate other plants than bee a. In a nested matrix, we find, for an arbitrary pair of bee species, that the bee species with less than or an equal number of pollinated plants always pollinates a subset of the plant species that is pollinated from the other one. Hence, it cannot happen that a specialized bee pollinates other plants than a bee which pollinates more plant species. An opposite scenario is that all bee pairs pollinate pairwise different plants like in matrix
M of Example 1.
Example 1. Matrix is not nested since each bee pollinates a different plant than every other bee.
Imagine the extinction of a specialized bee in a nested matrix. Since this bee only pollinates plants, which are also pollinated from more generalized bees (bees which pollinate more plants), the pollination for all plants is still preserved. Even the extinction of a generalized bee would preserve the pollination of plants that are still pollinated by specialized bees. In contrast, the extinction of bee a in matrix
M of Example 1 would lead to the extinction of plants
A and
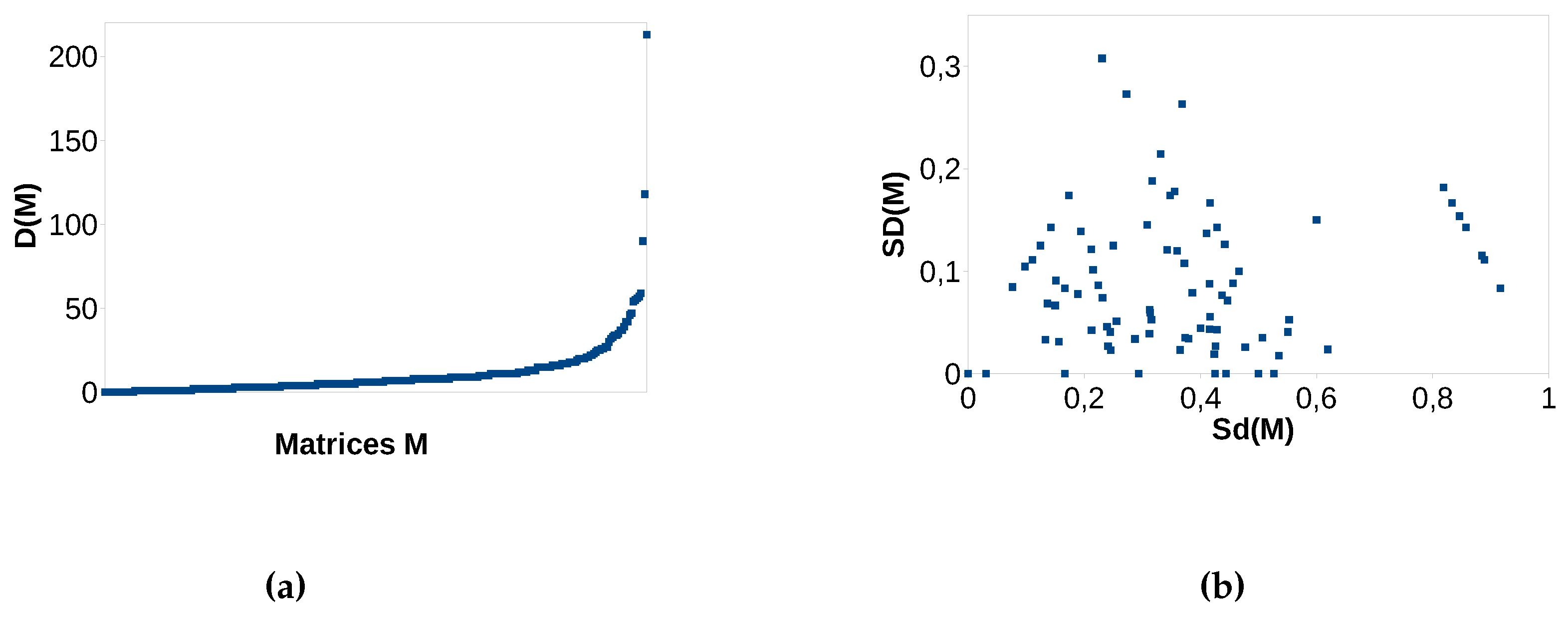
D because no other bee can provide this service for these plants. Real data matrices are mostly not nested (which can be seen in our experimental
Section 3). However, it seems that understanding “how much” ecological matrices are nested is important due to the fact that nestedness has been suggested as key to the stability of complex ecological systems. More information about stability and nestedness can be found, for example, in the following articles [
3,
4,
5,
6].
Notice that it is always possible to exchange columns and rows in a given matrix such that one finds non-increasing column and row sums. This indeed results in a different matrix. Anyway, it contains exactly the same relationships as before, and let us consider Example 2.
Example 2. Exchanging columns 2 and 3 in matrix N of Figure 1 leads to matrix which does not fulfill the property of non-increasing column sums. Moreover, in this matrix, it is more difficult to recognize nestedness because rows 2, 3 and 4 do not consist of consecutive 1
s followed by consecutive 0
s. However, if columns and rows are ordered in regards to non-increasing row and column sums, it is easy to decide whether a given matrix is nested or not. We give a mathematical definition for nestedness.
Definition 1 (nestedness)
. Let M be an -matrix where the columns and rows are sorted such that we find for the column sums , and for the row sums Then, matrix M is callednestedif and only if each of the m rows start with consecutive 1 s followed by consecutive 0 s.
In graph theory, a nested matrix is usually referred to as
Ferrers matrix [
7]. We will use equivalently the two terms. Moreover, a whole theory around Ferrers matrices exists, and it is worth reconsidering theoretical insights regarding the nestedness metrics. Let
M be an
-matrix with non-increasing row sums
and non-increasing column sums
The corresponding
Ferrers matrixF is the matrix where each row
i of
F starts with
1 s followed by
0 s. It can be achieved in several
shifts. A shift is the movement of a 1 in a row from a right to a left column with entry
i.e., it is an exchange of entries
and
for
leading to the matrix
with
,
and
for all index pairs of
M with
or
The corresponding Ferrers matrix of matrix
M in Example 1 can be seen in Example 3.
Example 3. Matrix is the corresponding Ferrers matrix of matrix M in Example 1. It can be achieved by shifting 1 s to the left. Matrix F has the same row sums like M but different column sums.
Example 3 shows that Ferrers matrix F has the same row sums as M but different column sums. They can be calculated in counting for each row i in M the number of rows which have at least row sum
Remark 1. Let M be an -matrix M where the columns and rows are sorted such that we find for the column sums , and for the row sums Furthermore, let F be its corresponding Ferrers matrix. Then, Matrix F also has row sums but different column sums, i.e., where
Remark 1 gives row and column sums for a corresponding Ferrers matrix
F of
The vector
of column sums is also known as
conjugate vector of
Furthermore, it is a classical result that there is exactly one matrix with these column and row sums [
8] (
Section 2).
Remark 2. The corresponding Ferrers matrix F of a matrix M is unique.
Ferrers matrices together with
majorisation theory are classical concepts for deciding the question: whether a matrix
M (or its bipartite graph) exists with given row and column sums. When matrix
M is interpreted as the bi-adjacency matrix, then the corresponding graph of matrix
F (considered now as bi-adjacency matrix) is then a
threshold graph. For an overview about these problems, we recommend Brualdi’s book [
9] or Mahadev’s and Peled’s book [
10].
In most real world data, a matrix
M is not nested. Hence, the natural question arises of “how far the ecological matrix is from being nested”. Brualdi and Sanderson answered this question with the metrics of
discrepancy [
11]. (In ecology, it is common to use the notion BR instead of
[
2]). It is based on the fact that every matrix
M has a unique Ferrers matrix
F that can be achieved by shifting 1 s in each row from larger to smaller column indices with entry 0 (see Remark 2). The discrepancy of a given matrix
M is the minimum number of shifts of 1 s to the left required to achieve its Ferrers matrix. A value of 0 represents a nested matrix. This means that the
the larger the discrepancy is (the more shifts are needed to yield the Ferrers matrix),
the less a matrix is nested. Hence, the discrepancy metrics measures the strength to be non-nested. Consider matrix
M in Example 4 and its unique Ferrers matrix
F. In the first row, it is possible to shift the 1 in the third column to the second column, and the 1 of the fourth column to the third one. However, one only needs a single shift, i.e., moving the 1 of the fourth column to the second one. Following this approach for each row, we find
The discrepancy can be easily calculated. However, there is a conceptual problem when applied to an ecological data set. Exchanging two different columns
i and
in
holding the same column sums, leads to another matrix
with
The values of the discrepancy for
M and
can be different. However, both bi-adjacency matrices represent exactly the same relationships between bees and plants. Moreover, these bi-adjacency matrices correspond to two different labelled bipartite graphs. (A graph is labelled if all vertices in vertex set
V are indexed by a unique number, i.e.,
V consists of vertices
where
) Their structure, however, is exactly the same. Formally, those graphs are called
isomorphic. (Two labelled graphs
and
are called isomorphic if there exists a permutation
of vertex labels such that edge
if and only if
.) Consider Example 4.
Example 4. Bi-adjacency matrices and represent two isomorphic bipartite graphs, i.e., both graphs look the same but plants C and D have in M column indices 3 and 4, whereas in matrix plant C and D are labelled with 4 and Moreover, in M, we need to shift the four bold 1 s to achieve its corresponding Ferrers matrix , i.e., In matrix , we only need to shift three 1 s to achieve F leading to
For ecological purposes, these labels are not necessary. They are only assigned to columns and rows due to the mathematical denotation of columns and rows. The value of the discrepancy is dependent on a “random” order of bees and pollinators in a data set, and its resulting order of columns and rows in its bi-adjacency matrix.
We propose an extended metric based on the discrepancy to repair this problem. We define for a matrix M set which contains all matrices that can be achieved by permuting in M columns and rows with identical sums. (Notice, if we consider all matrices in as bi-adjacency matrices, then the corresponding set of bipartite graphs is isomorphic. Consider a bipartite graph in this set. The definition of isomorphic graphs allows only the permutation of vertex labels with the same vertex degree in U or This corresponds to the permutation of columns and rows in the corresponding bi-adjacency matrix with the same sums.) However, changing the row order in M would not change the discrepancy of M because the number of minimal shifts in each row stays the same. Finding an optimal labelling for a given bipartite graph such that its bi-adjacency matrix has a minimum discrepancy under all bi-adjacency matrices of isomorphic graphs of can be done by only permuting the columns of its bi-adjacency matrix. We define the isomorphic discrepancy of M as the minimum number of shifts under all matrices in to achieve F. In other words, the isomorphic discrepancy finds an optimal labelling for a bipartite graph regarding a minimum number of shifts in its bi-adjacency matrix to achieve its nested matrix In Example 4, the discrepancy of M would be overestimated or the nestedness be underestimated, respectively.
We show that the isomorphic discrepancy for a matrix
M can be calculated in transforming our problem into less than
nminimum perfect weighted matching problems in a bipartite graph. (Given a bipartite graph,
with
and weight function
The
perfect matching problem asks for a subset
of edges such that each vertex of
G is in exactly one edge of
The
minimum perfect weighted matching problem asks for a perfect matching
M minimizing
) More clearly, we construct
for each fixed column sum s a bipartite graph
separately. Each edge
in a minimum perfect weighted matching of
tells us that column
i in matrix
M needs to be moved on position
In the resulting matrix, all columns with sum
s are reordered without violating the non-increasing column sums property. We repeat this approach whenever there exist at least two columns with the same column sum. The resulting matrix is a matrix in
with a minimum discrepancy. One of the most important books about matching theory is from Lovász and Plummer [
12]. One suitable algorithm for our scenario is from Gabow [
13], which achieves an
asymptotic running time, if we follow our approach.
Please observe that there exist other approaches and assumptions of what a nested matrix
N for
M has to look like. One approach is to find an
such that a minimal number of 0 s can be replaced by 1 s to become nested. This problem is known as the
minimal chain completion problem and was shown to be NP-complete in 1981 by Yannakakis [
14]. (It is an open problem whether there is an algorithm to find a solution in polynomial asymptotic running time for NP-complete problems. For more information consider the classical book of Garey and Johnson [
15].) The opposite problem is to delete a minimal number of 0s in
to achieve a nested matrix that is also NP-complete (exchange the rules of 1 s and 0 s to see this). An approximation algorithm was given in the Phd-thesis of Juntilla in 2011 [
16]. There are different definitions of nestedness in the ecological community, and several procedures to compute it. For an overview, see the nestedness guide [
2] which also includes the discrepancy of Brualdi and Sanderson (there called BR). In the next section, we give a formal definition of
isomorphic discrepancy, and show how it can be calculated in asymptotic running time
2. The Discrepancy Problem for Isomorphic Matrices
Let
M be an
bi-adjacency matrix with non-increasing row sums
, and non-increasing column sums
and
G the corresponding bipartite graph. Furthermore, let
be the set of all isomorphic graphs of
Then, all bi-adjacency matrices corresponding to a graph
can be achieved by permuting columns and rows with equal sums in
M. We denote the set of these matrices by
The discrepancy of
M given by Brualdi and Sanderson [
17] is the minimum number of shifts to achieve the corresponding Ferrers matrix
F as described in the last section. We denote the discrepancy of
M by
. We now give the definition of the
isomorphic discrepancy of M, which is defined by
In other words, we try to find a matrix in with minimal discrepancy. We also want to mention that this new metric should be invariant against the transposition of This can be achieved by transposing M to its transposed matrix and determining The general isomorphic discrepancy of M is then the minimum value (or the mean value) of and We here focus on because the calculation of can be done analogously.
A simple approach is to permute all columns and rows with equal sums, and to choose a permutation of
M that yields the minimum discrepancy. This can lead to the determination of exponentially many possible permutations. Please observe that it is sufficient for our problem to permute columns in
M and keep the rows fixed. The reason is that exchanging the order of rows with the same sums leads to exactly the same discrepancy because the corresponding rows of the Ferrers matrix are identical. Consider our Example 4. It can be seen that each permutation of the first three rows in
M or
lead to the same kind of shifts, and so to the same discrepancy as before. Following this approach, we will not necessarily generate all matrices in
but the subset
that consists of all matrices that were yielded by a permutation of columns in
M with the same sum. Hence, matrices in
possess the whole range of possible discrepancies that occur for matrices in
Hence,
it is sufficient to permute columns for the calculation of More formally, we find
Consider all matrices with row sums
and column sums
Then, it follows that all of them possess the same unique Ferrers matrix
F with column sums
(Recall that the lists are non-increasing.) Brualdi and Shen showed [
11] that there is always one matrix
A with row sums
and column sums
that has
minimum possible discrepancy , where
for
Matrix
A can be found by moving in
F 1 s from columns
j with
to columns
i with
Consider Example 5.
Example 5. For matrix we have , and corresponding Ferrers matrix We construct matrix with by shifting three 1 s of the second column in F to the fourth column. However, the minimum discrepancy under all isomorphic matrices of M is 4, i.e., This is true because each permutation σ of the last three columns of M leads to a matrix with A discrepancy of 3 cannot be achieved by a permutation of columns from
The set of all matrices with row sums and column sums can be partitioned in isomorphic matrix sets Example 5 shows that not all sets possess a matrix A with minimum possible discrepancy. If this had been the case, we would already know the value for each matrix. Unfortunately, it is not the case.
We want to develop a simple formula to calculate the discrepancy of a given matrix. We need this formula later to devise an approach for the isomorphic version. Notice that a shift in a matrix
M will always be applied when
and
Since
M and
F have identical row sums, and due to the construction of
F, there must always be an index
with
and
In matrices of our Examples 4 and 5, we marked these 0 s and 1 s. This means that the number of absences of 1 s in
when they are present in
correspond to the number of minimum shifts
. We define the difference of the
jth columns
and
of
-matrices
A and
by
Back to our problem, we find for all The reason is that a column has 1 s and a column has 1 s. This means that the minimum difference of both columns happens if all 1 s in also occur in . Then, Consider in Example 4 the third columns of M, and has all 1 s of F, whereas M has only two of them. Hence, but In cases where you find a 1 in , which does not occur in , this 1 needs to be shifted to a ’left’ column where F has a 1 and M a A 1 from a ’right’ column in M has to be shifted to to achieve the 1 s in F. In Example 4, this is the case for We put this connection in another formula for the discrepancy.
Proposition 1. Let M be an -matrix with non-increasing row sums and non-increasing column sums Let F be the corresponding Ferrers matrix. Then, the minimum number of shifts to achieve F from M can be calculated by .
Proof. The proof is by induction on the number
n of columns. For one column, we have
and so
and
We assume that
holds for all matrices with
m rows and
columns (induction hypothesis IH). Let us now consider a matrix an
matrix
Notice that
for each matrix
M and its corresponding Ferrers matrix
F due to the construction of
In column
of
M, we consider the set
I of all indices
i where
and
Due to the construction of a Ferrers matrix, and since
F and
M have equal row sums, we find for all
that
and
We construct matrix
B by exchanging, for all indices
, all entries
to 1, and all corresponding entries
to
Basically, we apply
shifts from the
nth column to the
th column. In matrix
B, we only find shiftable 1 s in smaller columns than in column
n due to our construction. We delete the last column of
B and get the
matrix
We apply the induction hypothesis on
Since (a) matrices
B and
have exactly the same number of minimal shifts (because in the first
columns they are completely identical, and
B has in the
nth column no shiftable 1 s due to our construction), and (b)
M has
more shifts (in the
nth column) than
and (c) matrices
M and
B are identical in the first
columns, and
M has in the
th column
more shifts, we get
☐
We denote an element of
by
where
is a permutation of the columns of
M that exchanges columns with same column sums. Recall (Equation (
2)) that this approach covers all possible discrepancies that exist in set
A permutation
can be divided in the convolution of
k permutations
if
M possesses
k different column sums, i.e.,
. Each
permutes all columns with column sum
independently, and keeps the indices of columns with different sums, i.e.,
We denote the set of all possible permutations
by
, and for all
by
For matrix
M in Example 4, we get six different matrices in set
. Each of them is built by a permutation
where
is the identity permutation which keeps the first column on index
and
permutes the second, third and fourth columns of
The same is true for
in Example 4. Notice that,
Let
be the number of columns with column sum
We construct
k different
sub-matrices
Each of them consists of all columns in
M with column sum
We denote the column indices
j in
by the same indices like in
i.e.,
The next Proposition states that calculating the minimum discrepancy of each matrix
can be done separately. The sum of all minimum discrepancies for each
is then the minimum discrepancy of
Proposition 2. Let M be an matrix M with non-increasing row sums and non-increasing column sums . Let F be the corresponding Ferrers matrix of M. Then, the isomorphic discrepancy of matrix M is Proof. Since the isomorphic discrepancy is the minimum discrepancy in the set of all matrices in
(Equation (
2)), and for each matrix
there exists a permutation
with
and
we get for the isomorphic discrepancy of
MSince each
is the convolution of
k permutations
that permute only the column indices with same sums (
), we can rewrite
Together with Equation (
4), and since
permutes columns with each fixed sum
independently, we get
☐
Proposition 2 states that we need to find for each sub-matrix
in
M with fixed column sums
a column order (a permutation
) that minimizes the following sum:
We transform the calculation of
for each sub-matrix (
) of
M to the determination of
minimum weighted perfect matching problem in a labelled
weighted complete bipartite graph (A bipartite graph
is called
complete if and only if each pair of vertices
with
and
is connected by an edge, i.e.,
If we have a weight function
which assigns to every edge in
G a weight, then
G is called
weighted ). In
, each vertex
corresponds to a column
of
where
Moreover, each vertex
corresponds to a column
of the Ferrers matrix
F of
M where
i.e., we consider the exactly same indices of columns in
F as in sub-matrix
For simplicity we denote vertices
by
, or,
by
, respectively. We assign each edge
the weight
. Now, we calculate a minimum weighted perfect matching
in each
Hence,
minimizes Equation (
5). Moreover, an edge
in
tells us that we have to put column
ℓ in
M on position
That is,
calculates how to permute the columns in
such that the discrepancy is minimal. Consider Example 6.
Example 6. For the -matrix with , we get -sub-matrix with column sums 2, and -matrix with column sums Hence, we have to construct two complete bipartite graphs and The Ferrers matrix for M is We get for vertices , , , and for vertices , , , . The edge weights between a vertex and are This leads in to a weight of 1 for all edges. Hence, every perfect matching in has weight , and is minimal. The order of the columns in does not matter. On the contrary, in , we find the minimum perfect weighted matching of weight Notice that and All other weights in are Hence, the perfect matching corresponds to the order of columns in M with weight , which is not the minimum. We need to exchange columns 3 and 4 in We get matrix with and Notice that corresponds to the value
Theorem 1. Calculating the isomorphic distance of an -matrix M needs asymptotic time.
Proof. Let
be a minimum weighted perfect matching in bipartite graph
for each
Then, permutation
with
for all edges
, and
for all
j with
is a permutation of the columns in sub-matrix
, which minimizes
in Equation (
5). With Proposition 2, and because the perfect matching has minimum weight, we get for
that
Let us now assume that each permutation that minimizes corresponds in each graph to a perfect matching with if for and This matching is by construction minimal.
Let
be the number of columns with sum
Then, the computation of a minimum weighted perfect matching in
is
(see, for example, [
13]). Since
(because
), we get a total asymptotic running time of
☐
Please observe that under certain conditions the construction of several graphs can be avoided. This happens for example when the corresponding Ferrers columns are completely equal like in Example 6 in . Then, all edge weights in are also equal, and therefore, each perfect matching is a solution.