An RNA Sequencing Transcriptome Analysis and Development of EST-SSR Markers in Chinese Hawthorn through Illumina Sequencing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and DNA Extraction
2.2. EST-SSR Mining from RNA-Seq and Primer Design
2.3. Primer Selection and PCR Amplification
2.4. Fluorescent-Labeled M13-SSR Markers and Capillary Electrophoresis
2.5. Data Analysis
2.6. Clustering Analysis
3. Results
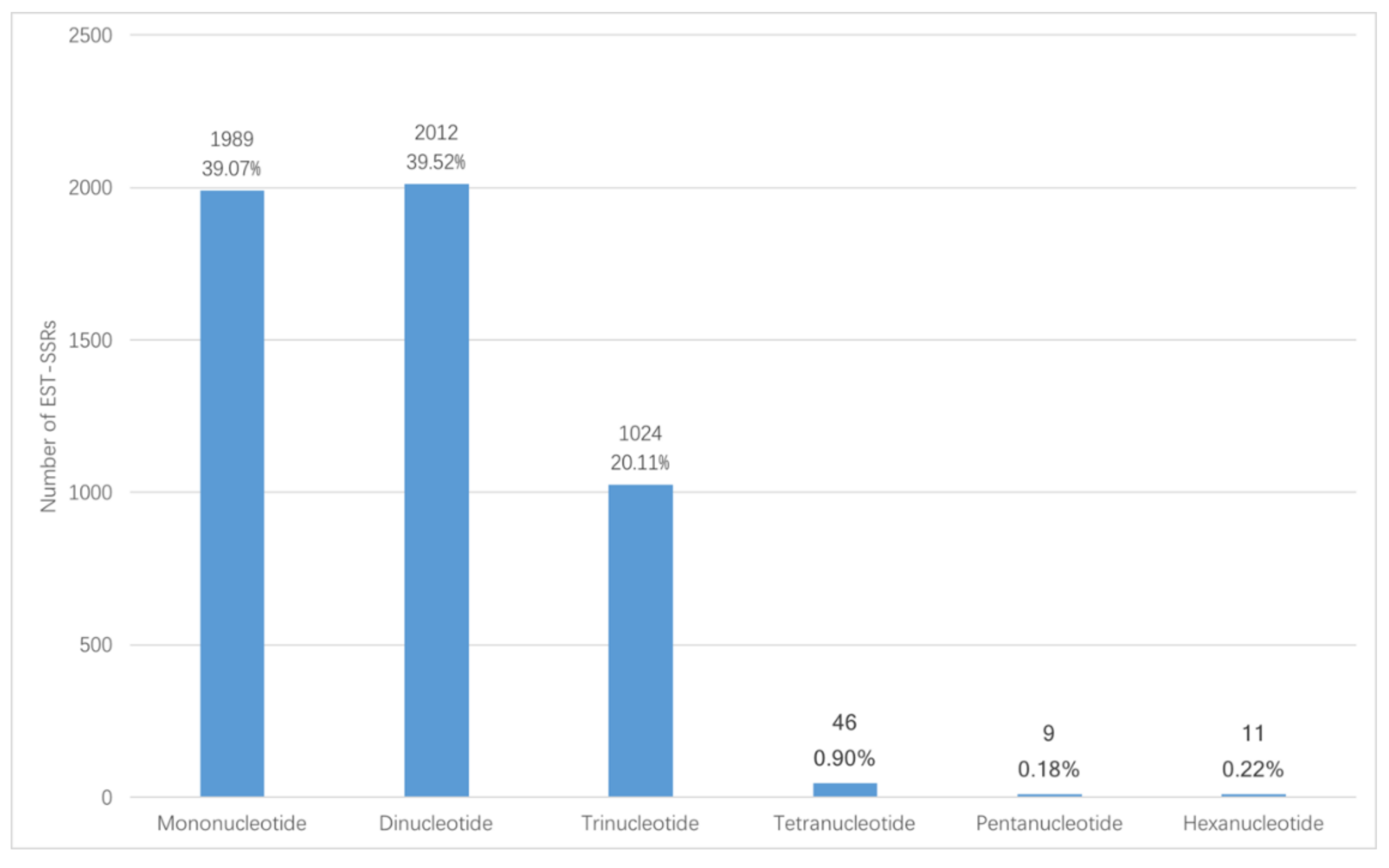
3.1. Frequency and Distribution of EST-SSRs
3.2. Development, Screening, and Polymorphic Validation of EST-SSRs
3.3. Genetic Diversity Analysis
3.4. Cluster Analysis Using EST-SSR Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Christensen, K.I. Revision of Crataegus sect. Crataegus and nothosect. Crataeguineae (Rosaceae-Maloideae) in the Old World. Syst. Bot. Monogr. 1992, 35, 1–199. [Google Scholar] [CrossRef]
- Brown, J.A.; Beatty, G.E.; Finlay, C.; Montgomery, I.; Tosh, D.G.; Provan, J. Genetic analyses reveal high levels of seed and pollen flow in hawthorn (Crataegus monogyna, Jacq.), a key component of hedgerows. Tree. Genet. Genomes 2016, 12, 58. [Google Scholar] [CrossRef]
- Betancourt-Olvera, M.; Nieto-Ángel, R.; Urbano, B.; González-Andrés, F. Analysis of the biodiversity of hawthorn (Crataegus spp.) from the morphological, molecular, and ethnobotanical approaches, and implications for genetic resource conservation in scenery of increasing cultivation: The case of Mexico. Genet. Resour. Crop. Evol. 2018, 65, 897–916. [Google Scholar] [CrossRef]
- Gundogdu, M.; Ozrenk, K.; Ercisli, S.; Kan, T.; Kodad, O.; Hegedus, A. Organic acids, sugars, vitamin C content and some pomological characteristics of eleven hawthorn species (Crataegus spp.) from Turkey. Biol. Res. 2014, 47, 21. [Google Scholar] [CrossRef] [PubMed]
- Rigelsky, J.M.; Sweet, B.V. Hawthorn: Pharmacology and therapeutic uses. Am. J. Health-Syst. Pharm. 2002, 59, 417. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Deng, J.; Wen, L.; You, L.; Zhao, Z.; Zhou, L. Release of phenolic compounds and antioxidant capacity of Chinese hawthorn “Crataegus pinnatifida” during, in vitro, digestion. J. Funct. Foods 2018, 40, 76–85. [Google Scholar] [CrossRef]
- Guo, T.J.; Jiao, P.J. Hawthorn (Crataegus) resources in China. HortScience 1995, 30, 1132–1134. [Google Scholar]
- Zhao, H.C.; Feng, B.T. China Fruit-Plant Monograph of Hawthorn (Crataegus) Flora; Zhongguo Linye Press: Beijing, China, 1996. [Google Scholar]
- Dai, H.; Zhang, Z.; Guo, X. Adventitious bud regeneration from leaf and cotyledon explants of Chinese hawthorn (Crataegus pinnatifida Bge. var major N.E.Br.). In Vitro Cell Dev. Biol. 2007, 43, 2–8. [Google Scholar] [CrossRef]
- Dai, H.; Guo, X.; Zhang, Y.; Li, Y.; Chang, L.; Zhang, Z. Application of random amplified polymorphic DNA and inter-simple sequence repeat markers in the genus Crataegus. Ann. Appl. Biol. 2009, 154, 175–181. [Google Scholar] [CrossRef]
- Dai, H.; Han, G.; Yan, Y.; Zhang, F.; Liu, Z.; Li, X.; Li, W.; Ma, Y.; Li, H.; Liu, Y.; et al. Transcript assembly and quantification by RNA-Seq reveals differentially expressed genes between soft-endocarp and hard-endocarp hawthorns. PLoS ONE 2013, 8, e72910. [Google Scholar] [CrossRef]
- Silfverberg-Dilworth, E.; Matasci, C.L.; Van de Weg, W.E.; Van Kaauwen, M.P.; Walser, M.; Kodde, L.P.; Soglio, V.; Gianfranceschi, L.; Durel, C.E.; Costa, F.; et al. Microsatellite markers spanning the apple (Malus x domestica Borkh.) genome. Tree. Genet. Genomes 2006, 2, 202–224. [Google Scholar] [CrossRef]
- Bao, L.; Chen, K.; Zhang, D.; Cao, Y.; Yamamoto, T.; Teng, Y. Genetic diversity and similarity of pear (Pyrus L.) cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. Genet. Resour. Crop. Evol. 2007, 54, 959. [Google Scholar] [CrossRef]
- Cao, Y.; Tian, L.; Gao, Y.; Liu, F. Genetic diversity of cultivated and wild Ussurian Pear (Pyrus ussuriensis Maxim.) in China evaluated with M13-tailed SSR markers. Genet. Resour. Crop. Evol. 2012, 59, 9–17. [Google Scholar] [CrossRef]
- Liang, C.; Wan, T.; Xu, S.; Li, B.; Li, X.; Feng, Y.; Cai, Y. Molecular identification and genetic analysis of cherry cultivars using capillary electrophoresis with fluorescence-labeled SSR markers. 3 Biotech 2018, 8, 16. [Google Scholar] [CrossRef]
- Dai, H. Molecular Identification and Enhancement of Germplasms in Hawthorn. Ph.D. Thesis, Shenyang Agricultural University, Shenyang, China, 2007. [Google Scholar]
- Ferrazzini, D.; Monteleone, I.; Belletti, P. Small-scale genetic diversity in oneseed hawthorn (Crataegus monogyna, Jacq.). Eur. J. Forest Res. 2008, 127, 407–414. [Google Scholar] [CrossRef]
- Rajeb, C.; Messaoud, C.; Chograni, H.; Bejaoui, A.; Boulila, A.; Rejeb, M.N.; Boussaid, M. Genetic diversity in Tunisian Crataegus azarolus L. var. aronia L. populations assessed using RAPD markers. Ann. Forest. Sci. 2010, 67, 512. [Google Scholar] [CrossRef]
- Yilmaz, K.U.; Yanar, M.; Ercislï, S.; Sahiner, H.; Taskin, T.; Zengin, Y. Genetic relationships among some hawthorn (Crataegus spp.) species and genotypes. Biochem. Genet. 2010, 48, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Serçe, S.; Şimşek, Ö.; Toplu, C.; Kamiloğlu, Ö.; Çalışkan, O.; Gündüz, K.; Özgen, M.; Kacar, Y.A. Relationships among Crataegus, accessions sampled from Hatay, Turkey, as assessed by fruit characteristics and RAPD. Genet. Resour. Crop. Evol. 2011, 58, 933–942. [Google Scholar] [CrossRef]
- Beigmohamadi, M.; Rahmani, F. Genetic variation in hawthorn (Crataegus spp.) using RAPD markers. Afr. J. Biotechnol. 2011, 10, 7131–7135. [Google Scholar] [CrossRef]
- Erfani-Moghadam, J.; Mozafari, M.; Fazeli, A. Genetic variation of some hawthorn species based on phenotypic characteristics and RAPD marker. Biotechnol. Biotechnol. Equip. 2016, 30, 247–253. [Google Scholar] [CrossRef]
- Zarei, A.; Erfanimoghadam, J.; Mozaffari, M. Phylogenetic analysis among some pome fruit trees of Rosaceae family using RAPD markers. Biotechnol. Biotechnol. Equip. 2017, 31, 247–253. [Google Scholar] [CrossRef]
- Han, X.Y.; Liang, Y.H.; Wang, Y.J.; Li, F.; Guo, T.; Xue, Y. Analysis of the origin and classification of C. brettschnederi by ISSR markers. J. Jilin Agric. Univ. 2009, 31, 164–167. [Google Scholar] [CrossRef]
- Mirali, N.; Alodat, M.; Haider, N.; Nabulsi, I. The genus Crataegus L.: An ecological and molecular study. Russ. J. Genet. 2011, 47, 26–34. [Google Scholar] [CrossRef]
- Rahmani, M.S.; Shabanian, N.; Khadivi-Khub, A.; Woeste, K.; Badakhshan, H.; Alikhani, L. Population structure and genotypic variation of Crataegus pontica inferred by molecular markers. Gene 2015, 572, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Sheng, F.; Chen, S.; Tian, J.; Li, P.; Qin, X.; Wang, L.; Li, J. Morphological and ISSR molecular markers reveal genetic diversity of wild hawthorns (Crataegus songorica K. Koch.) in Xinjiang, China. J. Integr. Agric. 2017, 16, 2482–2495. [Google Scholar] [CrossRef]
- Emami, A.; Shabanian, N.; Rahmani, M.S.; Khadivi, A.; Mohammad-Panah, N. Genetic characterization of the Crataegus, genus: Implications for in situ, conservation. Sci. Hortic. 2018, 231, 56–65. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, G.; Su, K.; Dong, W.; Guo, Y. Genetic Diversity of Hawthorn Germplasms Analyzed by SRAP Markers. Mol. Plant Breed. 2014, 12, 1281–1287. [Google Scholar] [CrossRef]
- Kalia, R.K.; Manoj, K.R.; Kalia, S.; Singh, R.; Dhawan, A. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Csencsics, D.; Brodbeck, S.; Holderegger, R. Cost-Effective, Species-Specific Microsatellite Development for the Endangered Dwarf Bulrush (Typha minima) Using Next-Generation Sequencing Technology. J. Hered. 2010, 101, 789–793. [Google Scholar] [CrossRef]
- Dickinson, T.A.; Lo, E.Y.Y.; Talent, N.; Love, R.M. Black-fruited hawthorns of western North America—One or more agamic complexes? Botany 2008, 86, 846–865. [Google Scholar] [CrossRef]
- Liebhard, R.; Gianfranceschi, L.; Koller, B.; Ryder, C.D.; Tarchini, R.; Van de Weg, E.; Gessler, C. Development and characterisation of 140 new microsatellites in apple (Malus domestica Borkh.). Mol. Breed. 2002, 10, 217–241. [Google Scholar] [CrossRef]
- Lo, E.Y.Y. Global and Fine Scale Molecular Studies of Polyploidy Evolution in Crataegus L. (Rosaceae). Ph.D. Thesis, University of Toronto, Toronto, ON, Canada, 2008. [Google Scholar]
- Lo, E.Y.Y.; Stefanovic, S.; Dickinson, T.A. Population genetic structure of diploid sexual and polyploid apomictic hawthorns (Crataegus; Rosaceae) in the Pacific Northwest. Mol. Ecol. 2009, 18, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Khiari, S.; Boussaid, M.; Messaoud, C. Genetic diversity and population structure in natural populations of Tunisian Azarole (Crataegus azarolus, L. var aronia L.) assessed by microsatellite markers. Biochem. Syst. Ecol. 2015, 59, 264–270. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, H.Y.; Zhang, Q.J.; He, L.I.; Zhang, Z.H. Assessment of genetic relationship in Crataegus genus by the apple SSR primers. J. Fruit Sci. 2008, 25, 521–525. [Google Scholar] [CrossRef]
- Taheri, S.; Lee Abdullah, T.; Yusop, M.; Hanafi, M.; Sahebi, M.; Azizi, P.; Shamshiri, R. Mining and Development of Novel SSR Markers Using Next Generation Sequencing (NGS) Data in Plants. Molecules 2018, 23. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Simsek, O.; Donmez, D.; Kacar, Y.A. RNA-Seq Analysis in Fruit Science: A Review. Am. J. Plant Biol. 2017, 2, 1–7. [Google Scholar] [CrossRef]
- Yang, M.; Dong, Z.; Cao, Q.; Chen, M. Transcriptomics analysis of Chinese hawthorn (Crataegus pinnatifida) provides insights into the biosynthesis of polyphenolic compounds. Plant Omics 2015, 8, 89–95. [Google Scholar]
- Xu, J.; Zhao, Y.; Zhang, X.; Zhang, L.; Hou, Y.; Dong, W. Transcriptome Analysis and Ultrastructure Observation Reveal that Hawthorn Fruit Softening Is due to Cellulose/Hemicellulose Degradation. Front. Plant. Sci. 2016, 7, R106. [Google Scholar] [CrossRef]
- Xing, W.; Liao, J.; Cai, M.; Xia, Q.; Liu, Y.; Zeng, W.; Jin, X. De novo assembly of transcriptome from Rhododendron latoucheae, Franch. using Illumina sequencing and development of new EST-SSR markers for genetic diversity analysis in Rhododendron. Tree. Genet. Genomes 2017, 13, 53. [Google Scholar] [CrossRef]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current trends in microsatellite genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef]
- Oetting, W.S.; Lee, H.K.; Flanders, D.J.; Wiesner, G.L.; Sellers, T.A.; King, R.A. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics 1995, 30, 450–458. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Barkley, N.A.; Dean, R.E.; Pittman, R.N.; Wang, M.L.; Holbrook, C.C.; Pederson, G.A. Genetic diversity of cultivated and wild-type peanuts evaluated with M13-tailed SSR markers and sequencing. Genet. Res. 2007, 89, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Jin, H.; Han, R.; Wang, Y.; Zhu, S. Fingerprinting and identification of closely related wheat (Triticum aestivum L.) cultivars using ISSR and fluorescence-labeled TP-M13-SSR markers. Aust. J. Crop. Sci. 2011, 5, 846. [Google Scholar]
- Jia, H.M.; Shen, Y.T.; Jiao, Y.; Wang, G.Y.; Dong, X.; Jia, H.J.; Du, F.; Liang, S.M.; Zhou, C.C.; Mao, W.H.; et al. Development of 107 SSR markers from whole genome shotgun sequences of Chinese bayberry (Myrica rubra) and their application in seedling identification. J. Zhejiang Univ-Sci. B. 2014, 15, 997. [Google Scholar] [CrossRef]
- Schlautman, B.; Covarrubias-Pazaran, G.; Fajardo, D.; Steffan, S.A.; Zalapa, J.E. Discriminating power of microsatellites in cranberry organelles for taxonomic studies in Vaccinium, and Ericaceae. Genet. Resour. Crop. Evol. 2016, 64, 1–16. [Google Scholar] [CrossRef]
- Du, F.; Wu, Y.; Zhang, L.; Li, X.W.; Zhao, X.Y.; Wang, W.H.; Gao, Z.S.; Xia, Y.P. De Novo Assembled Transcriptome Analysis and SSR Marker Development of a Mixture of Six Tissues from Lilium, Oriental Hybrid ‘Sorbonne’. Plant. Mol. Biol. Rep. 2015, 33, 281–293. [Google Scholar] [CrossRef]
- Ouyang, P.; Kang, D.; Mo, X.; Tian, E.; Hu, Y.; Huang, R. Development and Characterization of High-Throughput EST-Based SSR Markers for Pogostemon cablin Using Transcriptome Sequencing. Molecules 2018, 23, 2014. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.F.; Zhang, Z.H.; Dai, H.Y.; Zhang, Y.; Chang, L.L. Genetic Relationships of Some Hawthorns (Crataegus spp.) Derived from cp DNA PCR-RFLP. J. Shenyang Agric. Univ. 2008, 39, 664–668. [Google Scholar]
- Albarouki, E.; Peterson, A. Molecular and morphological characterization of Crataegus L. species (Rosaceae) in southern Syria. Bot. J. Linn. Soc. 2007, 153, 255–263. [Google Scholar] [CrossRef]
- Hodel, R.G.; Gitzendanner, M.A.; Germain-Aubrey, C.C.; Liu, X.; Crowl, A.A.; Sun, M.; Landis, J.B.; Segovia-Salcedo, M.C.; Douglas, N.A.; Chen, S.; et al. A New Resource for the Development of SSR Markers: Millions of Loci from a Thousand Plant Transcriptomes. Appl. Plant Sci. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Vaiman, D.; Mercier, D.; Moazami-Goudarzi, K.; Eggen, A.; Ciampolini, R.; Lépingle, A.; Velmala, R.; Kaukinen, J.; Varvio, S.L.; Martin, P.; et al. A set of 99 cattle microsatellites: Characterization, synteny mapping, and polymorphism. Mamm. Genome 1994, 5, 288–297. [Google Scholar] [CrossRef] [PubMed]
- Grohme, M.A.; Soler, R.F.; Wink, M.; Frohme, M. Microsatellite marker discovery using single molecule real-time circular consensus sequencing on the Pacific Biosciences RS. BioTechniques 2013, 55, 253–256. [Google Scholar] [CrossRef] [PubMed]



| Parameters Used in Screening | Data Generated by MIcroSAtellite (MISA) |
|---|---|
| Total number of sequences examined | 14,364 |
| Total size of examined sequences (bp) | 28,888,844 |
| Total number of identified SSRs | 5091 |
| Number of SSR-containing sequences | 4011 |
| Number of sequences containing more than one SSR | 863 |
| Number of SSRs present in compound formation | 273 |
| Primer Pairs | Forward Sequence (5′-3′) Reverse Sequence (5′-3′) | Motif Repeat | Product Size (bp) | Tm (°C) | Na | Ne | I | Ho | He | PIC |
|---|---|---|---|---|---|---|---|---|---|---|
| Pr1 | AATATTTGACCCGCTGCAAG TTCTGCAGGAAAAACCCATC | (AC)8 | 130 | 53.5 | 3 | 2.933 | 1.087 | 0.900 | 0.659 | 0.585 |
| Pr19 | ATCAGTTTCCATCCCTGTCG TGGTGGACGTAACAGCACTC | (TA)6 | 206 | 56.5 | 6 | 2.632 | 1.168 | 0.797 | 0.620 | 0.554 |
| Pr26 | GCTGCTGCAGTAAGCAATGA GCAACCCACAAAACTGGAAT | (CCT)6 | 103 | 54.5 | 6 | 2.271 | 0.986 | 0.913 | 0.560 | 0.469 |
| Pr29 | ATGAGGCTGACGAGAGAGGA TTTGCAGAACCCAAAAGTCC | (AG)7 | 235 | 55.5 | 4 | 1.407 | 0.585 | 0.214 | 0.289 | 0.272 |
| Pr31 | TTACACTCGCCAGAACCCTC CCGTCATGTTGAATCCTGTG | (CCG)5 | 182 | 56.5 | 3 | 2.010 | 0.790 | 0.729 | 0.503 | 0.407 |
| Pr56 | GACGAGAATAACTCGCCTCG TGCACGACTTGCAGTACCTC | (TCG)5 | 199 | 57.5 | 6 | 2.454 | 1.117 | 0.824 | 0.592 | 0.522 |
| Pr59 | CTGCAAGATTGGAGGAGGAG TCAAGGGGAGTGCTCTCAGT | (TGT)5 | 166 | 57.5 | 2 | 2.000 | 0.693 | 0.829 | 0.500 | 0.375 |
| Pr81 | AGGAAGAGGAGGAATAGCCG GGATTCGCAGAGGATGTTGT | (GA)7 | 157 | 56.5 | 4 | 2.181 | 0.873 | 0.754 | 0.542 | 0.441 |
| Pr83 | TTCCTTCTCACGCAAAATCC TGGGTTTTGGAAGCTTTGAG | (CT)6 | 116 | 53.5 | 4 | 1.280 | 0.457 | 0.194 | 0.219 | 0.206 |
| Pr100 | GGACACCTTCTTTGGCACTC TGTGGGTTGTGTGTTTTGCT | (CT)9 | 173 | 55.5 | 4 | 1.479 | 0.561 | 0.250 | 0.324 | 0.280 |
| Pr110 | CGCCGTAAACAGAGAGAGGA GTCGGAGAAAATGGTGTCGT | (CT)8 | 144 | 56.0 | 4 | 2.537 | 1.071 | 0.841 | 0.606 | 0.533 |
| Pr114 | TTCAACCTCCATCCATCCAT CAAGCCTCATCAGAACACGA | (GCC)6 | 123 | 54.0 | 5 | 2.340 | 0.976 | 0.855 | 0.573 | 0.480 |
| Pr117 | CTCAATGCAGTGGGAACTCA AGAGCTTGAGCAAGCAGAGG | (AG)6 | 262 | 56.0 | 2 | 1.258 | 0.359 | 0.232 | 0.205 | 0.184 |
| Pr125 | GATGTGGACGATTGAGTTGC GCATGTAGCCCACAAGACAA | (TG)6 | 266 | 54.5 | 3 | 1.540 | 0.582 | 0.414 | 0.351 | 0.298 |
| Pr134 | GGGTTGGTGAAAGCCCTAAT ATGCATACGCAGCAGTCTTG | (ATC)6 | 130 | 56.0 | 4 | 2.654 | 1.054 | 0.814 | 0.623 | 0.548 |
| Pr146 | AGAAGATGACGACCACGACC TGCGATTCGAAACCCTAATC | (TTC)6 | 269 | 54.5 | 4 | 2.373 | 1.008 | 0.786 | 0.579 | 0.518 |
| Pr152 | CGCTTGGTTAACTGTGGAGC TTGGACTCCACACATGCAAT | (AC)6 | 127 | 56.0 | 4 | 2.133 | 0.880 | 0.714 | 0.531 | 0.444 |
| Pr167 | AACACTTCCCTCACCGTCAC TAGATTGCCAGGACCAGACC | (GTC)5 | 209 | 57.0 | 6 | 2.090 | 0.878 | 0.754 | 0.521 | 0.425 |
| Pr171 | CATACCCACTCAAGGGCTGT GGCCTTTGACTCCAAATGAA | (CA)8 | 226 | 56.0 | 14 | 3.214 | 1.571 | 0.857 | 0.689 | 0.642 |
| Pr174 | TCTTTCTCACCGAACCCATC TCGTAATCGGCGAGAGAGTT | (CT)8 | 227 | 54.5 | 4 | 1.773 | 0.785 | 0.132 | 0.436 | 0.389 |
| Pr175 | TTGCACTTGTGGCTAGATCG CAATTCATTGCACTTCGGTG | (AT)7 | 133 | 53.5 | 9 | 3.010 | 1.383 | 0.870 | 0.668 | 0.616 |
| Pr205 | ACCGTAGGAAGTCCAACACG CCCTCAGTTCAAGGAATACTCG | (GAG)5 | 125 | 57.5 | 4 | 2.510 | 1.016 | 0.899 | 0.602 | 0.523 |
| Pr211 | TGTTGCTGCAATCTGCTTTC CCACTTGGAGGCAGTAGTGA | (AAT)6 | 183 | 56.0 | 4 | 3.082 | 1.228 | 0.900 | 0.676 | 0.615 |
| Pr224 | AAACTTCATCGGTCCGTCAG CTCTTCCTTGCTGCATCCTC | (AG)6 | 242 | 56.0 | 10 | 2.456 | 1.207 | 0.836 | 0.593 | 0.527 |
| Pr226 | ACGAGAGCATGCAGGAGAAT GCTTTCCGAACCTTCAACAG | (GGC)5 | 242 | 56.0 | 4 | 2.007 | 0.755 | 0.829 | 0.502 | 0.390 |
| Pr235 | GCAAACAATGGGCACACATA GCTGTGCATGAGATGGAAGA | (GAG)5 | 163 | 54.5 | 9 | 2.563 | 1.191 | 1.000 | 0.610 | 0.533 |
| Pr237 | CGGGATCAAGCAATGAAGAT CAGAGCCATGGTTCAGGAAT | (CCT)5 | 242 | 54.5 | 2 | 2.000 | 0.693 | 1.000 | 0.500 | 0.375 |
| Pr239 | ACCATGGAAGATTCGTGGAG CCAATGAGCAAAAGCGTGTA | (CTT)5 | 196 | 54.5 | 4 | 2.196 | 0.902 | 0.729 | 0.545 | 0.456 |
| Pr244 | GCCGACTCTACTTGAAGGGA TACGGATAAACCCAGCTTCG | (GTT)6 | 173 | 56.0 | 2 | 1.998 | 0.693 | 0.657 | 0.500 | 0.375 |
| Pr246 | GCAAGGGACTTGAAATTGGA ATCCGATCGGTTCTGTGAAG | (GA)9 | 174 | 54.5 | 13 | 2.259 | 1.397 | 0.609 | 0.557 | 0.542 |
| Pr251 | TCTTCCTTCTCCGCTCTCTG CCCATTTCTGTCTGCTCCAT | (AG)9 | 227 | 56.0 | 5 | 2.771 | 1.112 | 0.838 | 0.639 | 0.569 |
| Pr255 | GCCGTATGGAGGGTCCTTAG GCGGAGGTGATTGGAGACTA | (TC)6 | 177 | 58.5 | 2 | 1.845 | 0.651 | 0.710 | 0.458 | 0.353 |
| Pr295 | GCAATCGATGCATACAGCTC ACAAGAGTGTCAAGGCCCAG | (AG)9 | 225 | 56.0 | 3 | 2.047 | 0.772 | 0.729 | 0.512 | 0.401 |
| Mean | / | / | / | / | 4.939 | 2.221 | 0.924 | 0.709 | 0.524 | 0.450 |
| Standard Error | / | / | / | / | 0.515 | 0.088 | 0.049 | 0.042 | 0.022 | 0.118 |
| Source | Dai et al., 2013 [11] | Yang et al., 2015 [42] | Xu et al., 2016 [43] | |
|---|---|---|---|---|
| Materials | C. pinnatifida H8 | C. pinnatifida S7 | C. pinnatifida “Zezhouhong” | C. pinnatifida “Qiujinxing” and “Ruanroushanlihong3Hao” |
| Clean reads | 11,538,395 | 14,659,624 | 32,332,207 | 23,183,612 * |
| Total bases (bp) | 2,140,589,552 | 2,743,145,672 | 6,530,452,306 | 5,839,454,759 * |
| Number of >200 bp contigs | 460,119 | 515,118 | / | 2,182,914 |
| Mean length of contigs (bp) | 130 | 131 | / | 77 |
| Number of >200 bp transcripts | 54,662 | 62,653 | 83,817 | 199,204 |
| Mean length of transcripts (bp) | 756 | 846 | 938 | 1056 |
| N50 length of transcripts (bp) | 1237 | 1421 | 1615 | 1656 |
| Number of unigenes | 39,663 | 41,723 | / | 72,837 |
| Mean length of unigenes (bp) | 656 | 703 | / | 696 |
| Sequencer | Illumina HiSeq 2000 | Illumina HiSeq 2000 | Illumina HiSeq 2500 | |
| Source | Dai et al., 2013 [11] | Yang et al., 2015 [42] | Our Study | |||
|---|---|---|---|---|---|---|
| Mono-nucleotide | 992 | 31.25% | / | / | 1989 | 39.07% |
| Di-nucleotide | 1254 | 39.51% | 6707 | 64.05% | 2012 | 39.52% |
| Tri-nucleotide | 677 | 21.33% | 3118 | 29.77% | 1024 | 20.11% |
| Tetra-nucleotide | 29 | 0.91% | 159 | 1.52% | 46 | 0.90% |
| Penta-nucleotide | 4 | 0.13% | 156 | 1.49% | 9 | 0.18% |
| Hexa-nucleotide | 4 | 0.13% | 332 | 3.17% | 11 | 0.22% |
| Number of compound SSRs | 214 | 6.74% | / | / | 273 | 5.36% |
| Total number of identified SSRs | 3174 * | / | 10,472 | / | 5091 ** | / |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, S.; Dong, W.; Lyu, T.; Lyu, Y. An RNA Sequencing Transcriptome Analysis and Development of EST-SSR Markers in Chinese Hawthorn through Illumina Sequencing. Forests 2019, 10, 82. https://doi.org/10.3390/f10020082
Ma S, Dong W, Lyu T, Lyu Y. An RNA Sequencing Transcriptome Analysis and Development of EST-SSR Markers in Chinese Hawthorn through Illumina Sequencing. Forests. 2019; 10(2):82. https://doi.org/10.3390/f10020082
Chicago/Turabian StyleMa, Suliya, Wenxuan Dong, Tong Lyu, and Yingmin Lyu. 2019. "An RNA Sequencing Transcriptome Analysis and Development of EST-SSR Markers in Chinese Hawthorn through Illumina Sequencing" Forests 10, no. 2: 82. https://doi.org/10.3390/f10020082




